test case for very large runs #1963
JamiePringle
started this conversation in
General
Replies: 0 comments
Sign up for free
to join this conversation on GitHub.
Already have an account?
Sign in to comment
Uh oh!
There was an error while loading. Please reload this page.
Uh oh!
There was an error while loading. Please reload this page.
-
The attached code runs 30 days of particle releases in a global coastal ocean current field from the Mercator GLORYS V12R1 ocean circulation model from Copernicus Oceans. The problem is relatively large, with 1,549,325 particles released every day for 30 days for a total release of 46,479,750 particles.
The purposes of this code is to provide regression testing on large parcels runs, and to illustrate how to configure parcels for large and efficient runs. The timings given below might help folks understand what performance can be expected from parcels.
The files for the run can be found here and are:
The directories which need to be created (and mercatorDataFullGrid populated) are
The timings given below are the time it takes to run on a dual 16 core EPYC 7343 system with a relatively fast and cached ZFS file system. Most of these runs were made on a system which had recently accessed all of the data, so the data was mostly already in the large SSD based cache.
The timing are repeated for three different ways to partition the particles. It is clear that for large problems, choosing a good partitioning method can pay substantial dividends. Partitioning should optimally satisfy two constraints: 1) give the same amount of work to each MPI job, and 2) make the particles close in space so that a given MPI jobs will read in the same chunks of data from the circulation data. (It is important to get the chunking of the circulation data inputs and the float trajectory outputs configured appropriately before worrying about getting the partitioning optimal! See #1473 for discussions of output chunking and the parallel tutorial for discussions of input chunking.)
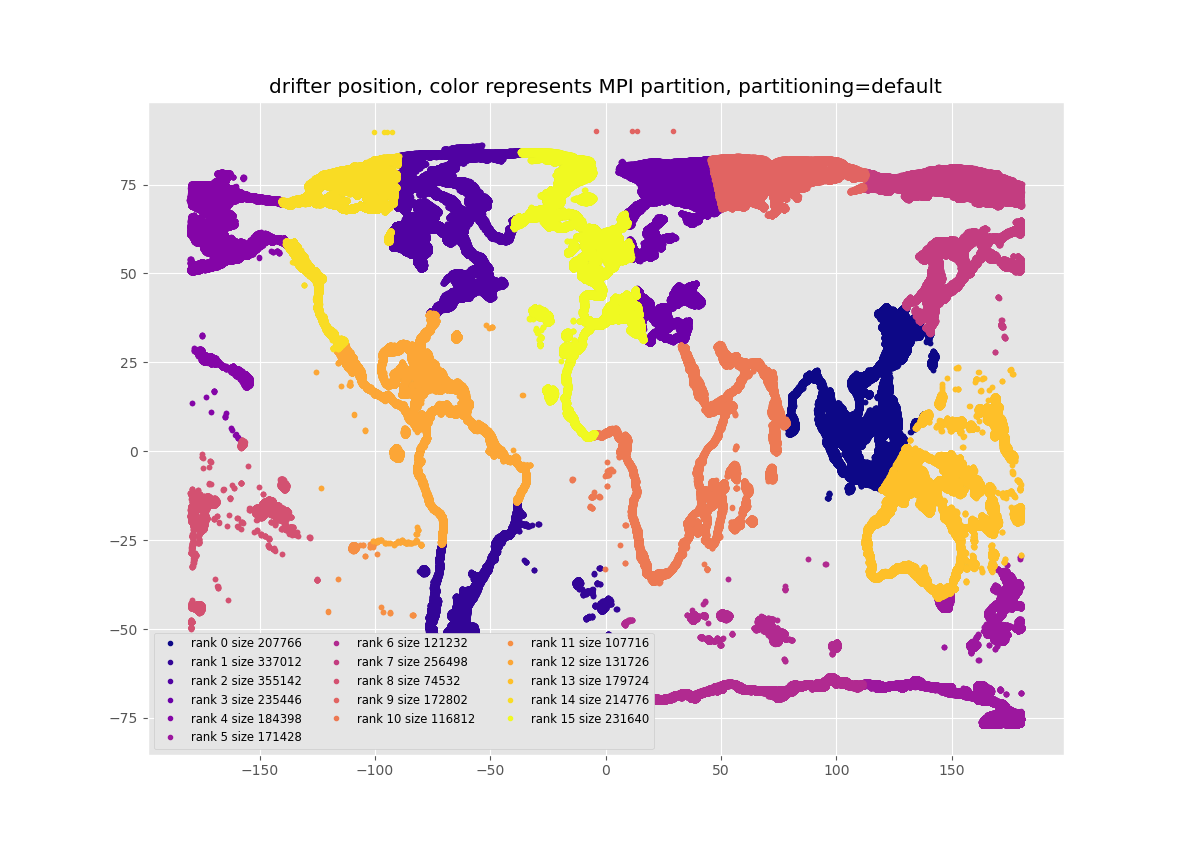
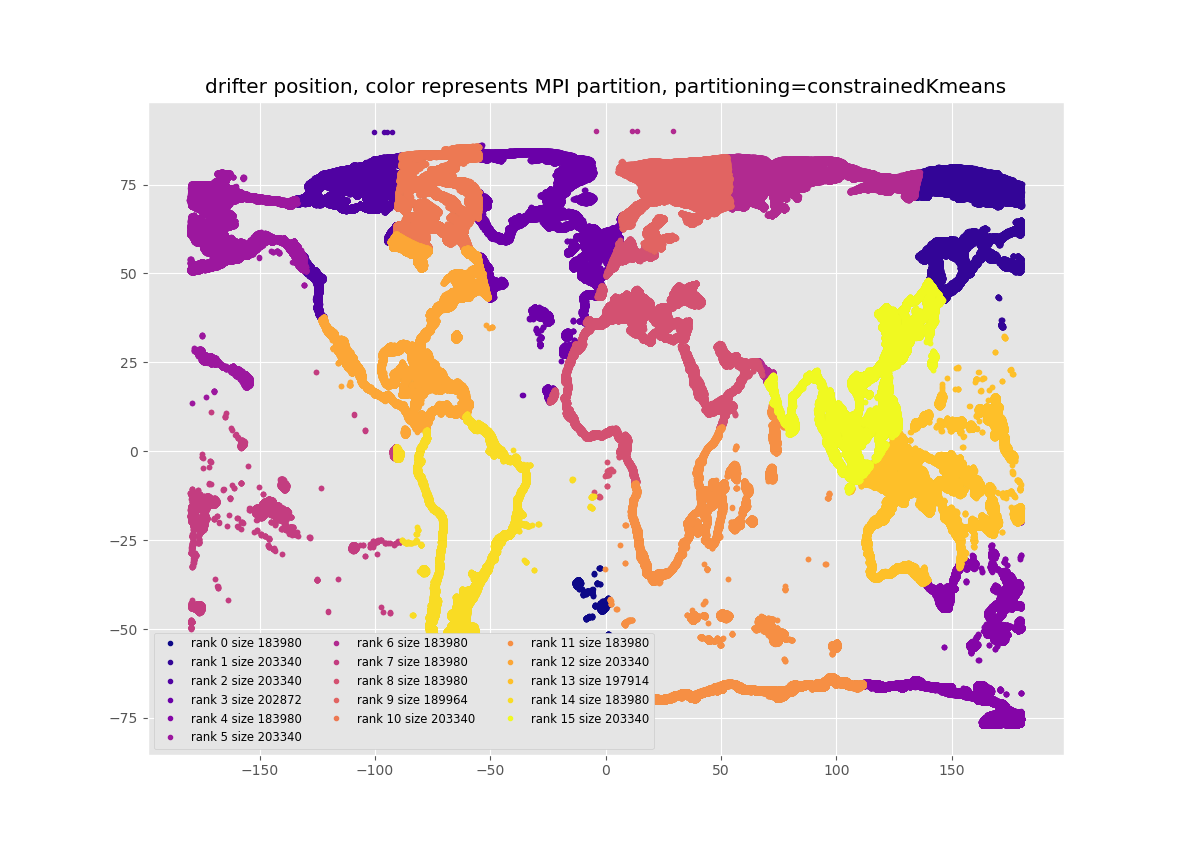
Here we examine 3 partitioning methods. See the figures at the end of this post. "Kmeans" is the default Kmeans based method of partitioning particles for MPI if SciKit-learn has been installed. "Uniform" just allocates the same number of particles to each core, in the order that they are listed in the list of the starting particle positions. There is no effort to make sure the particles are close to each other in space, though they mostly are in this list of starting positions. "Constrained-kMeans" uses a Kmeans algorithm which attempts to keep the same number of particles in each cluster, to within +/- 5% in this code. The underlying routine can be found here
Model run times
Serial time of 417.5 minutes
All times in minutes.
In the following figure, you can see the number of parcels runs/second as a function of the number of MPI processes. This would be linear in a perfect world -- twice the number of MPI processes would run the job twice as fast. The constrained-kmeans scales best, than uniform and Kmeans does the worst.

The consistent and reliable performance of the uniform initial partitioning of particles between MPI processes suggests that this might be a better default choice for partitioning than the default Kmeans scheme.
The reasons that the default "Kmeans" partitioning usually under-performs "uniform" and "constrained-kMeans" is that it usually puts different numbers of particles into each MPI process. The figures below show the partitioning and the number of particles in each partition, and it can be seen that this is most uneven with default Kmeans. For the default Kmeans, the smallest partition has 4.7 times fewer particles than the partition with the most particles. This uneven allocation of particles leads to inefficient allocation of effort, and slower run times. I think, but have not proven, that constrained-kMeans out-performs the "uniform" partitioning because it ensures that the particles in each cluster are closer together in Latitude/Longitude space, and this allows each MPI job to do less IO as each job needs to read fewer chunks of the circulation data.

Beta Was this translation helpful? Give feedback.
All reactions