In the following section we want to calculate the proportion of variation in species composition explained by each variable.Variation partitioning is a method of choice for the interpretation of beta diversity using tables of environmental and spatial variables. The technique of variation partitioning is used when two or more complementary sets of hypotheses can be invoked to explain the variation of an ecological response variable. A detailed description of variation partitioning can be found here.
rm(list = ls())
library(vegan)
library(plyr)
library(funrar)
library(ggplot2)
library(ggforce)
library(ggpubr)
OTU_Table = as.data.frame(read.csv2("../00_Data/05_Cercozoa_Seasonal_OTU_Table_min-freq-7633_transposed_withMetadata.csv",header = T))
#All
species = OTU_Table[,18:ncol(OTU_Table)]
species.mat = as.matrix(species)
species.mat = make_relative(species.mat)
SampleMetadata = as.data.frame(OTU_Table[,1:17])
#We need the Hellinger-transformed OTUs table with OTUs as columns and a table of metadata: Microhabitat, season and treespecies.
OTUCer.hel = decostand(species.mat, "hellinger")
#Ground
OTU_Ground <- OTU_Table[ which(OTU_Table$Stratum=='Ground'), ]
species.Ground = OTU_Ground[,18:ncol(OTU_Ground)]
species.mat.Ground = as.matrix(species.Ground)
species.mat.Ground = make_relative(species.mat.Ground)
SampleMetadata.Ground = as.data.frame(OTU_Ground[,1:17])
OTUCer.hel.Ground = decostand(species.mat.Ground, "hellinger")
#Canopy
OTU_Canopy <- OTU_Table[ which(OTU_Table$Stratum=='Canopy'), ]
species.Canopy = OTU_Canopy[,18:ncol(OTU_Canopy)]
species.mat.Canopy = as.matrix(species.Canopy)
species.mat.Canopy = make_relative(species.mat.Canopy)
SampleMetadata.Canopy = as.data.frame(OTU_Canopy[,1:17])
OTUCer.hel.Canopy = decostand(species.mat.Canopy, "hellinger")First we plot the variation partitioning of our variables with the varprat function within the vegan package. Variation partitioning (using RDA, CCA or db-RDA) among up to four matrices of environmental variables. First argument (Y) is dependent variable, usually the matrix of species composition (the function calculates RDA, or, if chisquare = TRUE, CCA), but could be also only a single variable (in that case it calculates linear regression) or distance matrix (applying db-RDA using the function capscale). Further arguments (up to four) are (groups of) explanatory variables. The function uses either formula interface (with ~, see examples) or matrices.
#All
Cervarp.all <- varpart(OTUCer.hel, ~ Season, ~ Microhabitat, ~TreeSpecies, data = SampleMetadata)
#Ground
Cervarp.Ground <- varpart(OTUCer.hel.Ground, ~ Season, ~ Microhabitat, ~TreeSpecies, data = SampleMetadata.Ground)
#Canopy
Cervarp.Canopy <- varpart(OTUCer.hel.Canopy, ~ Season, ~ Microhabitat, ~TreeSpecies, data = SampleMetadata.Canopy)#All
plot(Cervarp.all, digits = 1, Xnames = c('Season', 'Microhabitat', 'TreeSpecies'), bg = c('navy', 'tomato', 'green'))#Ground
plot(Cervarp.Ground, digits =1, Xnames = c('Season', 'Microhabitat', 'TreeSpecies'), bg = c('navy', 'tomato', 'green'))#Canopy
plot(Cervarp.Canopy, digits = 1, Xnames = c('Season', 'Microhabitat', 'TreeSpecies'), bg = c('navy', 'tomato', 'green'))Unfortunately it is not so easy to modify the plot afterwards. Consequently, we repeat the plotting with a dummy dataframe in ggplot to modify coloration and save settings.
#create dummy dataframe
df.venn.all <- data.frame(x = c(3, 1, 2),y = c(1, 1,2.8),labels = c('Microhabitat\n0.32', 'Season\n0.01',"Tree species\n0.03"))
#Ground
df.venn.Ground<- data.frame(x = c(3, 1, 2),y = c(1, 1,2.8),labels = c('Microhabitat\n0.31 **', 'Season\n0.05 **',"Tree species\n0.02 **"))
#Canopy
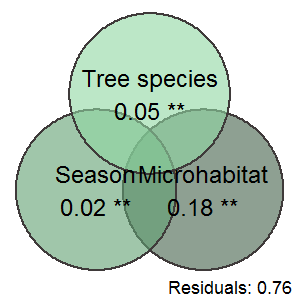
df.venn.Canopy<- data.frame(x = c(3, 1, 2),y = c(1, 1,2.8),labels = c('Microhabitat\n0.18 **', 'Season\n0.02 **',"Tree species\n0.05 **"))
#Plot
#All
Varpart <- ggplot(df.venn.all, aes(x0 = x, y0 = y, r = 1.5, fill = labels)) +
geom_circle(alpha = .6, size = 1, colour = "#3f3a3a",show.legend = FALSE) +
coord_fixed()+
annotate("text", x = df.venn.all$x , y = df.venn.all$y,label=df.venn.all$labels ,size = 6) +
scale_fill_manual(values = c("#660033","#B5A642","#094d35")) +
theme_void()
#annotate("text", x = 2 , y =1,label="A and B" ,size = 4) +
#annotate("text", x = 1.35 , y =2,label="B and C" ,size = 4) +
#annotate("text", x = 2.7 , y =2,label="A and C" ,size = 4) +
#annotate("text", x = 2 , y =1.6,label="A and B and C" ,size = 2)+theme_void()
Varpart#Ground
A <- ggplot(df.venn.Ground, aes(x0 = x, y0 = y, r = 1.5, fill = labels)) +
geom_circle(alpha = .6, size = 1, colour = "#3f3a3a",show.legend = FALSE) +
coord_fixed()+
annotate("text", x = df.venn.Ground$x , y = df.venn.Ground$y,label=df.venn.Ground$labels ,size = 6) +
# scale_fill_manual(values = c("#82662a","#bea773","#daba73")) +#old colors: brownish
scale_fill_manual(values = c("#52182b","#99597f","#822242")) +
theme_void()+
annotate("text", x = 3.5 , y =-0.8,label="Residuals: 0.65" ,size = 4.5) #+
#annotate("text", x = 2 , y =1,label="A and B" ,size = 4) +
#annotate("text", x = 1.35 , y =2,label="B and C" ,size = 4) +
#annotate("text", x = 2.7 , y =2,label="A and C" ,size = 4) +
#annotate("text", x = 2 , y =1.6,label="A and B and C" ,size = 2)+theme_void()
A#Canopy
B <- ggplot(df.venn.Canopy, aes(x0 = x, y0 = y, r = 1.5, fill = labels)) +
geom_circle(alpha = .6, size = 1, colour = "#3f3a3a",show.legend = FALSE) +
coord_fixed()+
annotate("text", x = df.venn.Canopy$x , y = df.venn.Canopy$y,label=df.venn.Canopy$labels ,size = 6) +
scale_fill_manual(values = c("#42614a","#61a072","#8fd7a2")) +
theme_void()+
annotate("text", x = 3.5 , y =-0.8,label="Residuals: 0.76" ,size = 4.5)
#annotate("text", x = 2 , y =1,label="A and B" ,size = 4) +
#annotate("text", x = 1.35 , y =2,label="B and C" ,size = 4) +
#annotate("text", x = 2.7 , y =2,label="A and C" ,size = 4) +
#annotate("text", x = 2 , y =1.6,label="A and B and C" ,size = 2)+theme_void()
Bcombi = ggarrange(A, B,
labels = c("A", "B"),
ncol = 2, nrow = 1, font.label = list(size = 16, color = "black"),
common.legend = T, legend = "bottom", align = "h")+
theme(plot.margin = margin(1,1,1,1, "cm"))
combi#save plot
#ggsave("VarPar_300_336x168.jpeg", plot = combi,
# device = "jpeg", dpi = 300, width = 336, height = 168,
# units = "mm")
#ggsave("VarPar_600_336x168.pdf", plot = combi,
# device = "pdf", dpi = 600, width = 336, height = 168,
# units = "mm")
#ggsave("VarPar_600_336x168.tif", plot = combi,
# device = "tiff", dpi = 600, width = 336, height = 168,
# units = "mm")
#ggsave("VarPar_600_336x168.png", plot = combi,
# device = "png", dpi = 600, width = 336, height = 168,
# units = "mm")To identify the explanatory variables that significantly explained variation in protistan communities, forward selection was performed using the ordistep function within the vegan package (1000 Monte Carlo permutations, alpha < 0.05). .
#Statistics: forward selection
#Ground
rda.all<-rda(OTUCer.hel.Ground~Season+Microhabitat+TreeSpecies, data=SampleMetadata.Ground)
rda.all
# Forward selection using vegan???s ordistep().
# *******************************************
# ->Hint: This function allows the use of factors. Options are also available for stepwise and backward selection of the explanatory variables.
step.forward <- ordistep(rda(OTUCer.hel.Ground ~ 1, data=SampleMetadata.Ground), scope=formula (rda.all), direction="forward", pstep=1000)
#Start: OTUCer.hel.Ground ~ 1
# Df AIC F Pr(>F)
#+ Microhabitat 1 -76.004 26.1939 0.005 **
#+ Season 1 -56.952 3.4393 0.010 **
#+ TreeSpecies 2 -54.181 1.3060 0.160
#---
#Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#Step: OTUCer.hel.Ground ~ Microhabitat
# Df AIC F Pr(>F)
#+ Season 1 -79.191 5.1488 0.005 **
#+ TreeSpecies 2 -75.755 1.8096 0.005 **
#---
#Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#Step: OTUCer.hel.Ground ~ Microhabitat + Season
# Df AIC F Pr(>F)
#+ TreeSpecies 2 -79.288 1.9484 0.005 **
#---
#Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#Step: OTUCer.hel.Ground ~ Microhabitat + Season + TreeSpecies
#Canopy
rda.all<-rda(OTUCer.hel.Canopy~Season+Microhabitat+TreeSpecies, data=SampleMetadata.Canopy)
rda.all
# Forward selection using vegan???s ordistep().
# *******************************************
# ->Hint: This function allows the use of factors. Options are also available for stepwise and backward selection of the explanatory variables.
step.forward <- ordistep(rda(OTUCer.hel.Canopy ~ 1, data=SampleMetadata.Canopy), scope=formula (rda.all), direction="forward", pstep=1000)
#Start: OTUCer.hel.Canopy ~ 1
# Df AIC F Pr(>F)
#+ Microhabitat 6 -286.40 9.0477 0.005 **
#+ TreeSpecies 2 -256.72 6.2777 0.005 **
#+ Season 1 -251.65 5.3221 0.005 **
#---
#Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#Step: OTUCer.hel.Canopy ~ Microhabitat
# Df AIC F Pr(>F)
#+ TreeSpecies 2 -298.02 7.7636 0.005 **
#+ Season 1 -291.16 6.6192 0.005 **
#---
#Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#Step: OTUCer.hel.Canopy ~ Microhabitat + TreeSpecies
# Df AIC F Pr(>F)
#+ Season 1 -303.21 6.9828 0.005 **
#---
#Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#Step: OTUCer.hel.Canopy ~ Microhabitat + TreeSpecies + Season