|
| 1 | +[](https://github.com/astrobatty/polyfitter/tree/master/examples) |
| 2 | +<!--- [](https://arxiv.org/abs/1909.00446) --> |
| 3 | + |
| 4 | +# polyfitter - Polynomial chain fitting and classification based on light curve morphology for binary stars. |
| 5 | + |
| 6 | +The mathematical background of polynomial chain fitting (polyfit) is published in [Prša et al.,2008,ApJ,687,542](https://ui.adsabs.harvard.edu/abs/2008ApJ...687..542P/abstract). |
| 7 | +The paper that related to this code can be found [here](https://ui.adsabs.harvard.edu/abs/maybeoneday). |
| 8 | +This code is built upon the original [polyfit](https://github.com/aprsa/polyfit). |
| 9 | + |
| 10 | +## Installation |
| 11 | + |
| 12 | +This code depends on the GNU Scientific Library (GSL)[https://www.gnu.org/software/gsl/]. Before installation, make sure it is properly installed, e.g. running the following, which should return the location of the library: |
| 13 | + |
| 14 | +```bash |
| 15 | +pkg-config --cflags --libs gsl |
| 16 | +``` |
| 17 | + |
| 18 | +To install the package clone this git and go the directory: |
| 19 | +```bash |
| 20 | +git clone https://github.com/astrobatty/polyfitter+git |
| 21 | + |
| 22 | +cd ./polyfitter |
| 23 | +``` |
| 24 | + |
| 25 | +As `polyfitter` is dependent on certain python package versions, the easiest way to install it is through creating a conda environment: |
| 26 | +```bash |
| 27 | +conda create -n polyfit python=3.7 scikit-learn=0.23.2 cython=0.29.20 |
| 28 | + |
| 29 | +conda activate polyfit |
| 30 | + |
| 31 | +python setup.py install |
| 32 | +``` |
| 33 | + |
| 34 | +If the code is not used, the environment can be deactivated: |
| 35 | +```bash |
| 36 | +conda deactivate |
| 37 | +``` |
| 38 | + |
| 39 | +If you want to access this environment from jupyter you can do the followings: |
| 40 | +```bash |
| 41 | +conda install -c anaconda ipykernel |
| 42 | + |
| 43 | +python -m ipykernel install --user --name=polyfit |
| 44 | +``` |
| 45 | + |
| 46 | +Then after restarting your jupyter you'll be able to select this kernel. |
| 47 | + |
| 48 | +## Example interactive usage |
| 49 | + |
| 50 | +To create your own photomery, you'll need a Target Pixel File, such as [this one.](https://github.com/zabop/autoeap/blob/master/examples/ktwo212466080-c17_lpd-targ.fits) |
| 51 | +Then, after starting Python, you can do: |
| 52 | +```python |
| 53 | +from polyfitter import Polyfitter |
| 54 | + |
| 55 | +import numpy as np |
| 56 | + |
| 57 | +# Parameters from OGLE database |
| 58 | +ID = 'OGLE-BLG-ECL-040474' |
| 59 | +P=1.8995918 |
| 60 | +t0=7000.90650 |
| 61 | + |
| 62 | +# Load light curve from OGLE database |
| 63 | +# This is in magnitude scale |
| 64 | +path_to_ogle = 'http://ogledb.astrouw.edu.pl/~ogle/OCVS/data/I/'+ID[-2:]+'/'+ID+'.dat' |
| 65 | +lc = np.loadtxt(path_to_ogle).T |
| 66 | + |
| 67 | +# For clarity |
| 68 | +time = lc[0] |
| 69 | +mag = lc[1] |
| 70 | +err = lc[2] |
| 71 | + |
| 72 | +# Create Polyfitter instance by setting the brightness scale of your data |
| 73 | +# Set "mag" or "flux" scale |
| 74 | +pf = Polyfitter(scale='mag') |
| 75 | + |
| 76 | +# Run polynomial chain fitting |
| 77 | +t0new, phase, polyfit, messages = pf.get_polyfit(time,mag,err,P,t0) |
| 78 | +``` |
| 79 | + |
| 80 | +Plotting our results gives: |
| 81 | +```python |
| 82 | +import matplotlib.pyplot as plt |
| 83 | + |
| 84 | +plt.errorbar((time-t0new)/P%1,mag,err,fmt='k.') |
| 85 | +plt.errorbar((time-t0new)/P%1-1,mag,err,fmt='k.') |
| 86 | +plt.plot(phase,polyfit,c='r',zorder=10) |
| 87 | +plt.plot(phase+1,polyfit,c='r',zorder=10) |
| 88 | +plt.xlabel('Phase') |
| 89 | +plt.ylabel('Magnitude') |
| 90 | +plt.xlim(-0.5,1) |
| 91 | +plt.gca().invert_yaxis() |
| 92 | +plt.show() |
| 93 | +``` |
| 94 | +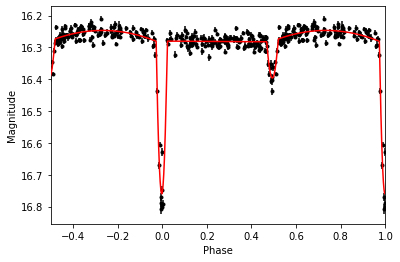 |
| 95 | + |
| 96 | +And the morphology classification: |
| 97 | +```python |
| 98 | +morp_array = pf.c |
| 99 | +print('Morphology type =' , morp_array[0] ) |
| 100 | +``` |
| 101 | + |
| 102 | +You can find a Google Colab friendly tutorial [in the examples](https://github.com/astrobatty/polyfitter/tree/master/examples/run_polyfit.ipynb). |
| 103 | + |
| 104 | +## Running as python script |
| 105 | + |
| 106 | +The difference from using the code interactively is that you have to put your code under a main function. See [here](https://github.com/astrobatty/polyfitter/tree/master/examples/run_polyfit_as_script.py) how to do it. |
| 107 | + |
| 108 | +## Available options |
| 109 | +- Polyfitter class instance: |
| 110 | + - `scale` The scale of the input data that will be used with this instance. Must be "mag" or "flux". |
| 111 | + - `debug` If `True` each fit will be displayed with auxiliary messages. |
| 112 | + |
| 113 | +- Getting polyfit: |
| 114 | + - `verbose` If `0` the fits will be done silently. Default is `1`. |
| 115 | + - `vertices` Number of equidistant vertices in the computed fit. Default is `1000`, which is mandatory to run the classification afterwards. |
| 116 | + - `maxiters` Maximum number of iterations. Default is `4000`. |
| 117 | + - `timeout` The time in seconds after a fit will be terminated. Default is `100`. |
| 118 | + |
| 119 | +## Contributing |
| 120 | +Feel free to open PR / Issue. |
| 121 | + |
| 122 | +## Citing |
| 123 | +If you find this code useful, please cite [X](https://ui.adsabs.harvard.edu/abs/maybeoneday), and [Prša et al.,2008,ApJ,687,542](https://ui.adsabs.harvard.edu/abs/2008ApJ...687..542P/abstract). Here are the BibTeX sources: |
| 124 | +``` |
| 125 | +@ARTICLE{2008ApJ...687..542P, |
| 126 | + author = {{Pr{\v{s}}a}, A. and {Guinan}, E.~F. and {Devinney}, E.~J. and {DeGeorge}, M. and {Bradstreet}, D.~H. and {Giammarco}, J.~M. and {Alcock}, C.~R. and {Engle}, S.~G.}, |
| 127 | + title = "{Artificial Intelligence Approach to the Determination of Physical Properties of Eclipsing Binaries. I. The EBAI Project}", |
| 128 | + journal = {\apj}, |
| 129 | + keywords = {methods: data analysis, methods: numerical, binaries: eclipsing, stars: fundamental parameters, Astrophysics}, |
| 130 | + year = 2008, |
| 131 | + month = nov, |
| 132 | + volume = {687}, |
| 133 | + number = {1}, |
| 134 | + pages = {542-565}, |
| 135 | + doi = {10.1086/591783}, |
| 136 | +archivePrefix = {arXiv}, |
| 137 | + eprint = {0807.1724}, |
| 138 | + primaryClass = {astro-ph}, |
| 139 | + adsurl = {https://ui.adsabs.harvard.edu/abs/2008ApJ...687..542P}, |
| 140 | + adsnote = {Provided by the SAO/NASA Astrophysics Data System} |
| 141 | +} |
| 142 | +``` |
| 143 | + |
| 144 | +## Acknowledgements |
| 145 | +This project was made possible by the funding provided by the Lendület Program of the Hungarian Academy of Sciences, project No. LP2018-7. |
0 commit comments