diff --git a/.github/workflows/docs.yml b/.github/workflows/docs.yml
index 4e1e663..a62bdde 100644
--- a/.github/workflows/docs.yml
+++ b/.github/workflows/docs.yml
@@ -11,25 +11,26 @@ jobs:
build:
runs-on: ubuntu-22.04
steps:
- - id: deployment
- uses: sphinx-notes/pages@v3
- with:
- publish: false
- - uses: peaceiris/actions-gh-pages@v4
- with:
- github_token: ${{ secrets.GITHUB_TOKEN }}
- destination_dir: ${{ github.ref_name }}
- publish_dir: ${{ steps.deployment.outputs.artifact }}
- uses: actions/checkout@v4
with:
- ref: ${{ github.event.repository.default_branch }}
+ ref: ${{ github.ref }}
+ path: ${{ github.ref_name }}
- uses: actions/checkout@v4
with:
ref: gh-pages
path: gh-pages
+ - name: Prepare Ruby and Jekyll
+ uses: ruby/setup-ruby@v1
+ with:
+ ruby-version: '3.3'
+ - name: Build deps and website
+ run: |
+ cd ${{ github.ref_name }}/docs/ \
+ && bundle install && bundle exec jekyll build
- name: Copy file
run: |
- cp ./assets/docs/index.html ./gh-pages/
+ mkdir -p ./gh-pages/${{ github.ref_name }}/ \
+ && cp -pr ${{ github.ref_name }}/docs/_site/* ./gh-pages/${{ github.ref_name }}/
- name: Deploy to GitHub Pages
uses: peaceiris/actions-gh-pages@v4
with:
diff --git a/docs/_config.yml b/docs/_config.yml
index da3939e..c7eba37 100644
--- a/docs/_config.yml
+++ b/docs/_config.yml
@@ -1,6 +1,7 @@
title: Master of Pores
description: Nextflow pipeline for analysis of Nanopore data from direct RNA sequencing.
-url: "https://biocorecrg.github.io/master_of_pores/"
+url: "https://biocorecrg.github.io"
+baseurl: "/master_of_pores/MOP1.0"
# Build settings
markdown: kramdown
diff --git a/docs/index.md b/docs/index.md
index 9d56ae4..b1aacbe 100644
--- a/docs/index.md
+++ b/docs/index.md
@@ -14,7 +14,7 @@ navigation: 1
-# 
+# 
# Nanopore analysis pipeline
Nextflow pipeline for analysis of Nanopore data from direct RNA sequencing. This is a joint project between [CRG bioinformatics core](https://biocore.crg.eu/) and [Epitranscriptomics and RNA Dynamics research group](https://www.crg.eu/en/programmes-groups/novoa-lab).
diff --git a/docs/nanomod.md b/docs/nanomod.md
index 7e1a126..fa82401 100644
--- a/docs/nanomod.md
+++ b/docs/nanomod.md
@@ -9,7 +9,7 @@ This module allows to predict the loci with RNA modifications starting from data
## Workflow
-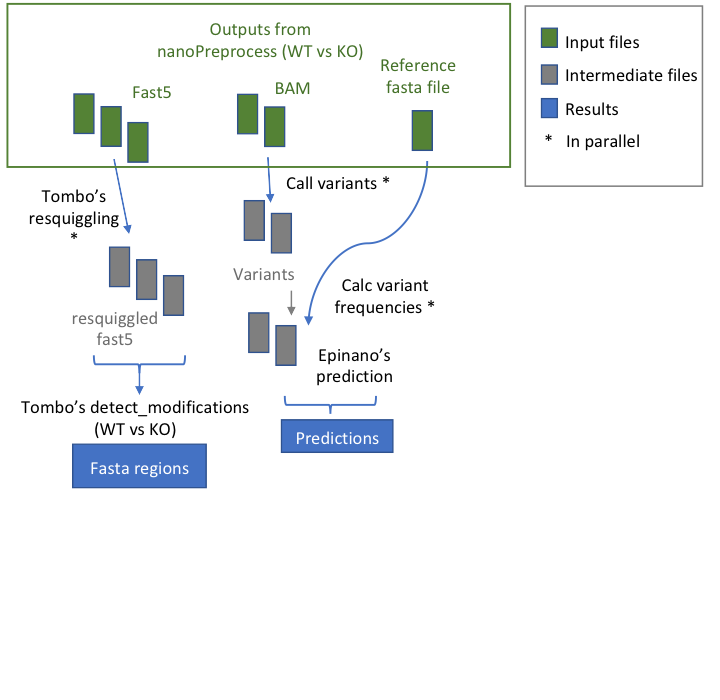 +
+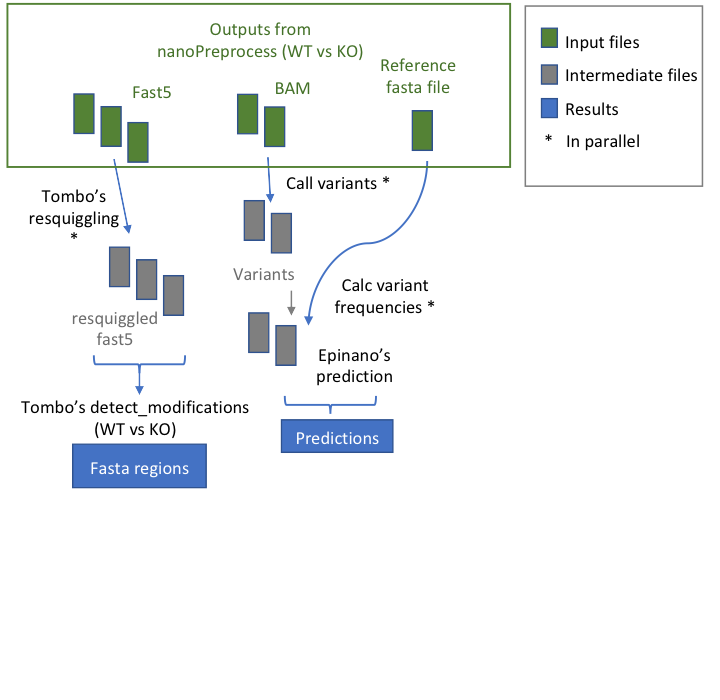 * **index_reference** index the reference file for Epinano
diff --git a/docs/nanopreprocess.md b/docs/nanopreprocess.md
index c310540..f90ce9d 100644
--- a/docs/nanopreprocess.md
+++ b/docs/nanopreprocess.md
@@ -11,7 +11,7 @@ This module takes as input the raw fast5 reads and produces a number of outputs
## Workflow
-
* **index_reference** index the reference file for Epinano
diff --git a/docs/nanopreprocess.md b/docs/nanopreprocess.md
index c310540..f90ce9d 100644
--- a/docs/nanopreprocess.md
+++ b/docs/nanopreprocess.md
@@ -11,7 +11,7 @@ This module takes as input the raw fast5 reads and produces a number of outputs
## Workflow
-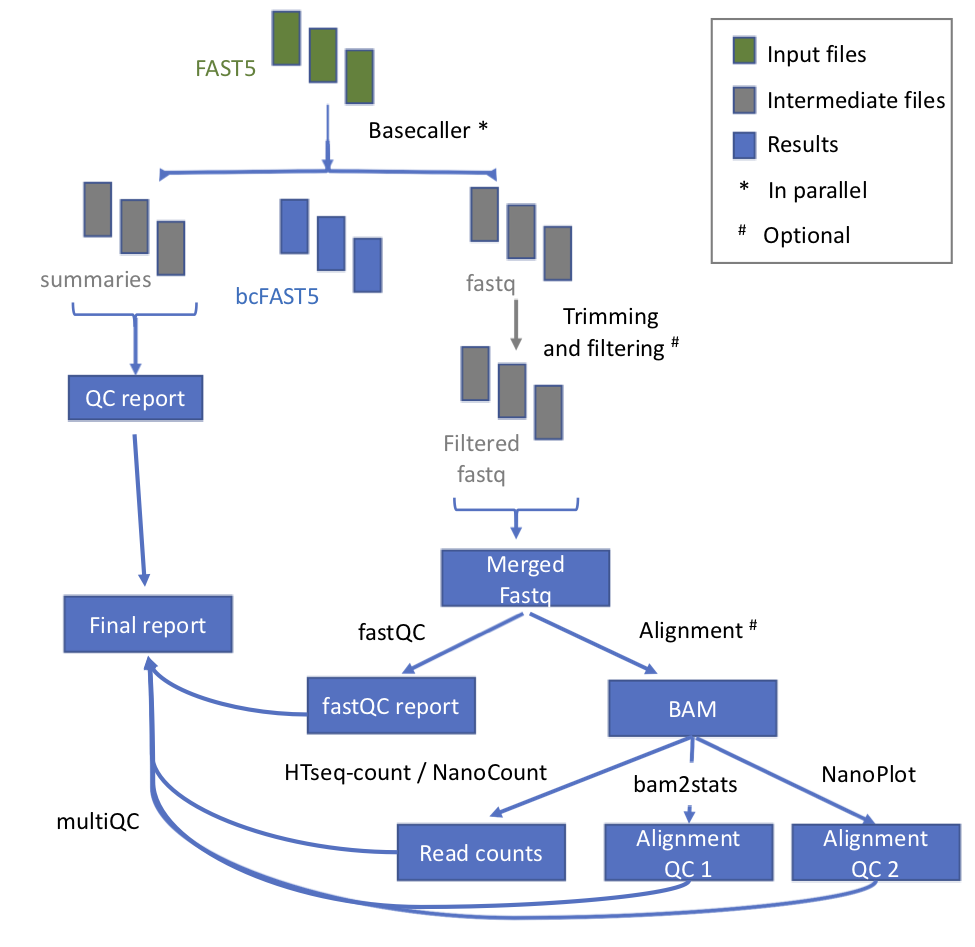 +
+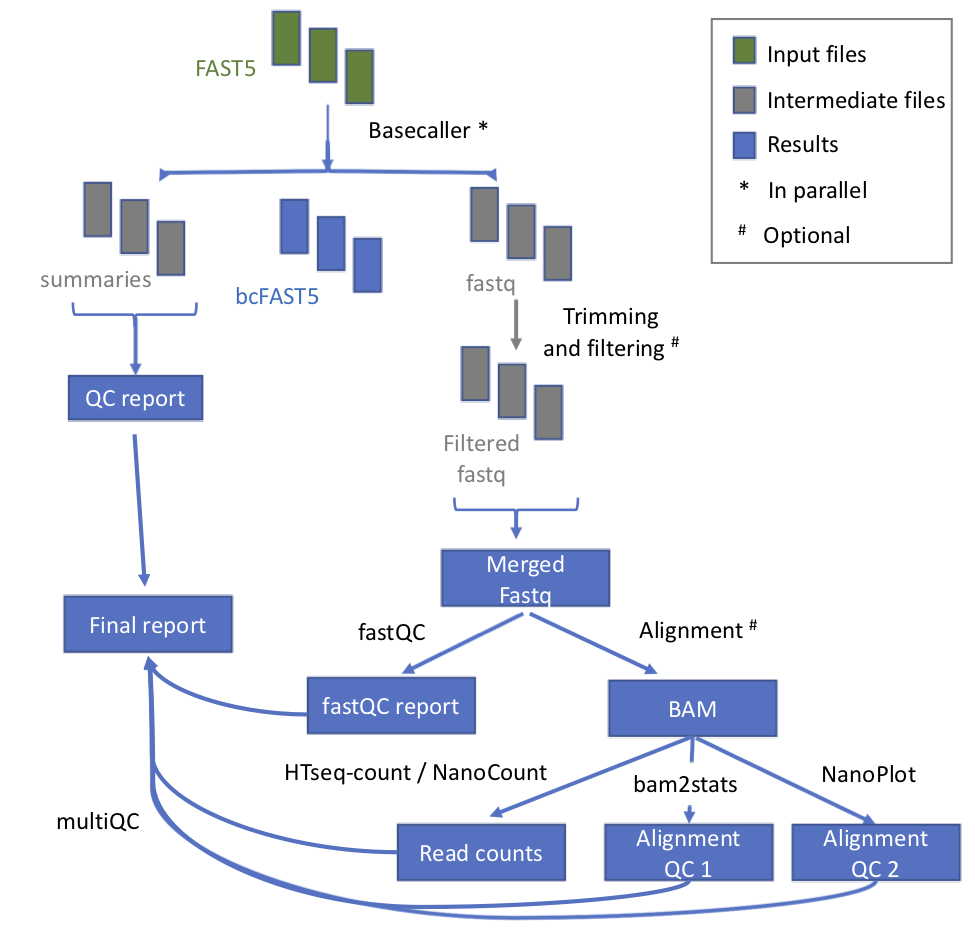 | Process name | Description |
| ------------- | ------------- |
diff --git a/docs/nanotail.md b/docs/nanotail.md
index d49f204..61fa708 100644
--- a/docs/nanotail.md
+++ b/docs/nanotail.md
@@ -9,7 +9,7 @@ This module allows to estimates polyA sizes by using two different methods (nano
# Workflow
-
| Process name | Description |
| ------------- | ------------- |
diff --git a/docs/nanotail.md b/docs/nanotail.md
index d49f204..61fa708 100644
--- a/docs/nanotail.md
+++ b/docs/nanotail.md
@@ -9,7 +9,7 @@ This module allows to estimates polyA sizes by using two different methods (nano
# Workflow
-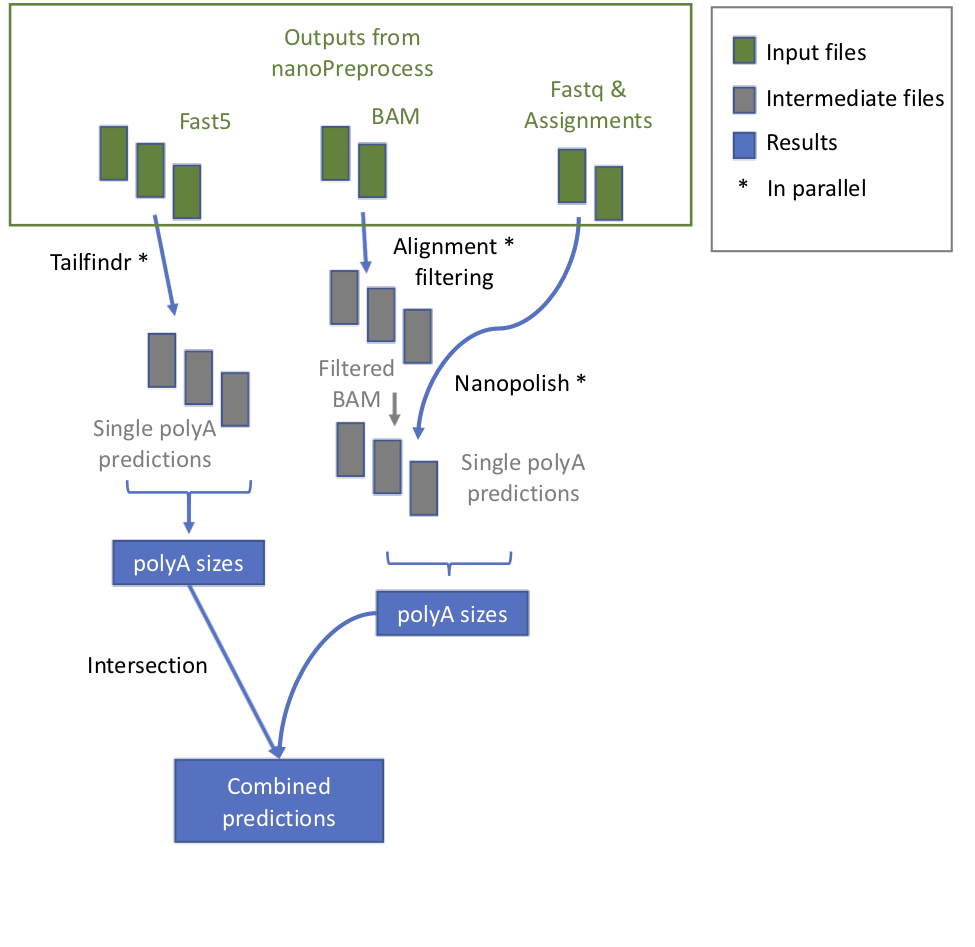 +
+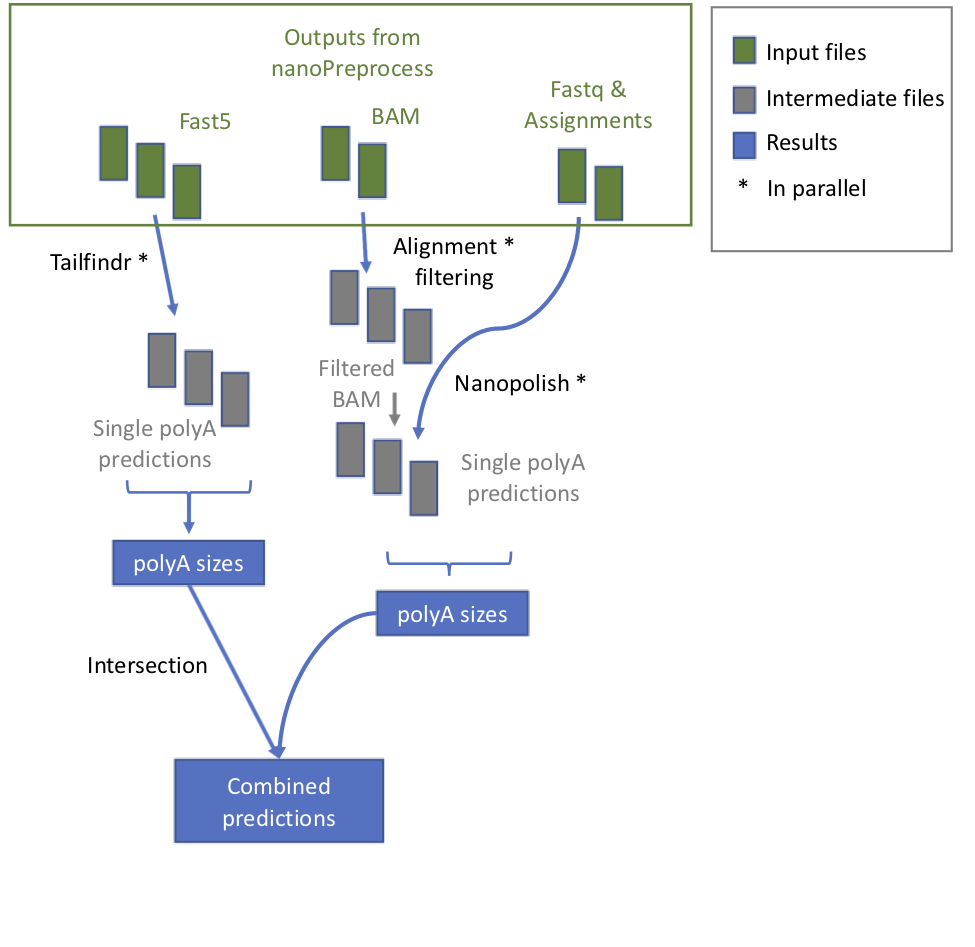 1. **check_reference** It verifies whether the reference is zipped and eventually unzip it
1. **tailfindr** it runs *tailfindr* tool in parallel.
1. **check_reference** It verifies whether the reference is zipped and eventually unzip it
1. **tailfindr** it runs *tailfindr* tool in parallel.
 +
+ * **index_reference** index the reference file for Epinano
diff --git a/docs/nanopreprocess.md b/docs/nanopreprocess.md
index c310540..f90ce9d 100644
--- a/docs/nanopreprocess.md
+++ b/docs/nanopreprocess.md
@@ -11,7 +11,7 @@ This module takes as input the raw fast5 reads and produces a number of outputs
## Workflow
-
* **index_reference** index the reference file for Epinano
diff --git a/docs/nanopreprocess.md b/docs/nanopreprocess.md
index c310540..f90ce9d 100644
--- a/docs/nanopreprocess.md
+++ b/docs/nanopreprocess.md
@@ -11,7 +11,7 @@ This module takes as input the raw fast5 reads and produces a number of outputs
## Workflow
- +
+ | Process name | Description |
| ------------- | ------------- |
diff --git a/docs/nanotail.md b/docs/nanotail.md
index d49f204..61fa708 100644
--- a/docs/nanotail.md
+++ b/docs/nanotail.md
@@ -9,7 +9,7 @@ This module allows to estimates polyA sizes by using two different methods (nano
# Workflow
-
| Process name | Description |
| ------------- | ------------- |
diff --git a/docs/nanotail.md b/docs/nanotail.md
index d49f204..61fa708 100644
--- a/docs/nanotail.md
+++ b/docs/nanotail.md
@@ -9,7 +9,7 @@ This module allows to estimates polyA sizes by using two different methods (nano
# Workflow
- +
+ 1. **check_reference** It verifies whether the reference is zipped and eventually unzip it
1. **tailfindr** it runs *tailfindr* tool in parallel.
1. **check_reference** It verifies whether the reference is zipped and eventually unzip it
1. **tailfindr** it runs *tailfindr* tool in parallel.