-
Notifications
You must be signed in to change notification settings - Fork 2
Open
Description
Super excited about the potential of rfsspec.
I tried it out with one of my HRRR kerchunk aggregations
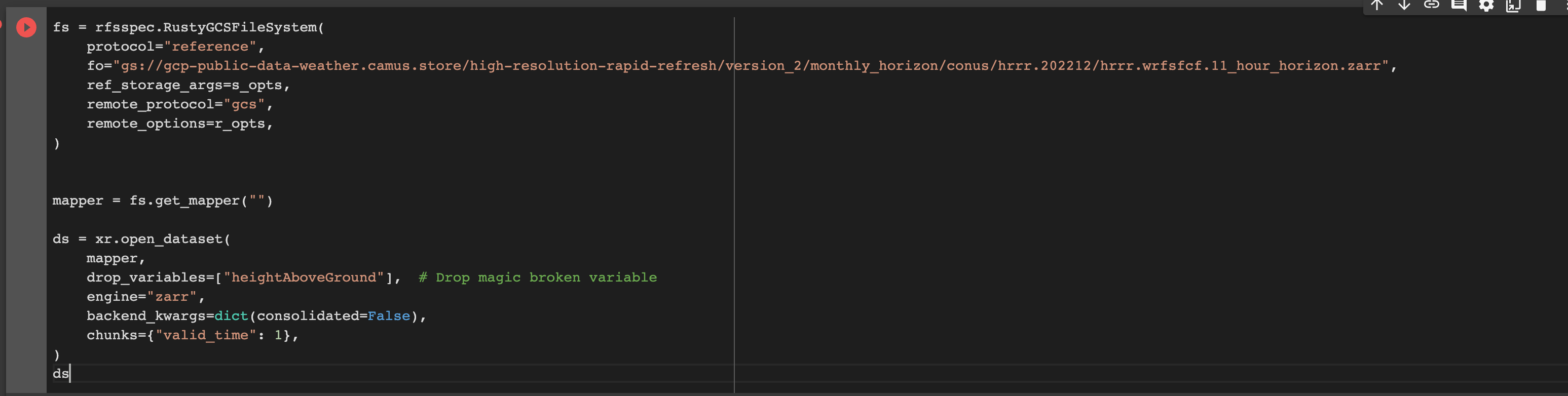
I get the following error:
File ~/.cache/bazel/_bazel_builder/99bd544777c52d5a0d8a66ea3b05626a/execroot/ritta/bazel-out/k8-dbg/bin/build/jupyter/jupyter.runfiles/common_deps_rfsspec/site-packages/rfsspec/gcs.py:41, in RustyGCSFileSystem.cat(self, path, recursive, on_error, start, end, **kwargs)
39 return out
40 else:
---> 41 return self.cat_file(paths[0], start=start, end=end, **kwargs)
File ~/.cache/bazel/_bazel_builder/99bd544777c52d5a0d8a66ea3b05626a/execroot/ritta/bazel-out/k8-dbg/bin/build/jupyter/jupyter.runfiles/common_deps_rfsspec/site-packages/rfsspec/gcs.py:28, in RustyGCSFileSystem.cat_file(self, url, start, end, **kwargs)
27 def cat_file(self, url, start=None, end=None, **kwargs):
---> 28 return gcs_cat_ranges([url], start=[start or 0], end=[end or 0], **self.kwargs)[0]
PanicException: called `Option::unwrap()` on a `None` value
Is this just a bug or a major missing feature in rfsspec to support reference file system in zarr?
Metadata
Metadata
Assignees
Labels
No labels