Description
Describe the bug
When I've tried to compute and discover motifs on my time series data, I noticed that the method mp.discover.motifs always returns only one motif and the same neighbor multiple times.
I've also tried to compute the motifs on the predefined "ecg-heartbeat-av" dataset by following exactly the steps as shown in this example of the documentary.
Unfortunately, I encounter the same problem and get results which are different from the example in the documentary.
Also, when I change the parameter k=3, I get the same motif multiple times. Furthermore, no matter which value I set for the parameter max_neighbor, I always get the same single neighbor that many times.
To Reproduce
Steps to reproduce the behavior:
Just follow the steps mentioned in the example from the documentary:
import matrixprofile as mp
ecg = mp.datasets.load('ecg-heartbeat-av')
ts = ecg['data']
profile = mp.compute(ts, windows=150)
profile = mp.discover.motifs(profile, k=1)
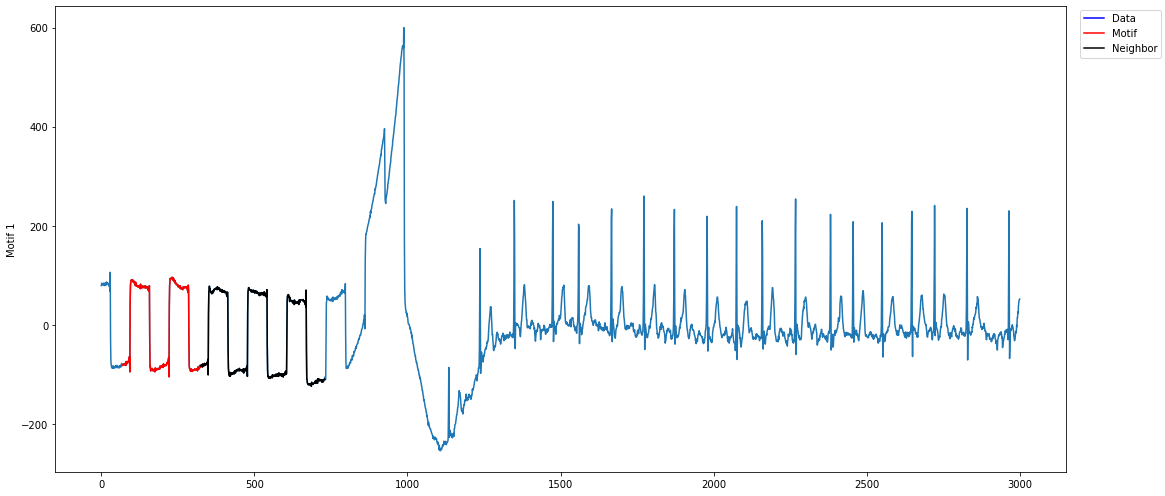
mp.visualize(profile)The motif plot I get looks like:

Note that I only get one neighbor, which is returned 10 times (default value of max_neighbors).
If I would change, for example, k=3, I get the same motif three times.
Expected behavior
The plot from the mentioned example:
As seen in the plot from the documentary, I would expect to get different neighbors. Also, I would expect that the method returns different motifs when the parameter k is increased.
Screenshots
See the plots above.
Desktop (please complete the following information):
- OS: Ubuntu LTS
- Python 3.8.10