|
1 | 1 | # Techniques for Machine Learning Applications |
2 | 2 |
|
3 | | -**Learning objectives:** |
| 3 | +**Learning Objectives:** |
4 | 4 |
|
5 | | -- THESE ARE NICE TO HAVE BUT NOT ABSOLUTELY NECESSARY |
| 5 | +- How to manipulate data through feature engineering\ |
| 6 | +- Select the most suitable model for your data\ |
| 7 | +- Learn about machine learning algorithms |
6 | 8 |
|
7 | | -## SLIDE 1 {-} |
| 9 | +## Goals of the Analysis and Nature of Data |
8 | 10 |
|
9 | | -- ADD SLIDES AS SECTIONS (`##`). |
10 | | -- TRY TO KEEP THEM RELATIVELY SLIDE-LIKE; THESE ARE NOTES, NOT THE BOOK ITSELF. |
| 11 | +### Output is *Continuous* |
11 | 12 |
|
12 | | -## Meeting Videos {-} |
| 13 | +- Example: How do explanatory variables such as lifestyle or chronic diagnoses affect LE / DALYs |
| 14 | +- Traditional **regression models**, including linear regression (OLS), ridge & lasso |
| 15 | +- Coefficient estimates quantify the association between changes in input and changes in outcome. |
13 | 16 |
|
14 | | -### Cohort 1 {-} |
| 17 | +### Output is *Categorical* or *Binary* |
| 18 | + |
| 19 | +- Outcome is categorical (e.g., disease/no disease) |
| 20 | +- Can use logistic regression (more common for *explaining*) |
| 21 | +- Or classification (more common for *predicting*) |
| 22 | + |
| 23 | +### Systemic Modelling / Simulation |
| 24 | + |
| 25 | +- For complex systems modelled by multiple equations |
| 26 | +- Typically more *predictive* |
| 27 | +- Have a series of equations to fit to data, for example SIR model |
| 28 | +- May wish to change parameters for sensitivity or explore how changes to inputs affects predicted outcome |
| 29 | + |
| 30 | +### Time-Series |
| 31 | + |
| 32 | +- Data has a temporal or seasonal aspect (influenza?) |
| 33 | +- Models like ARIMA can be used to model autocorrelation & trends |
| 34 | + |
| 35 | +## Statistical and Machine Learning Methods |
| 36 | + |
| 37 | +- Several pre-analysis steps are common to many methods |
| 38 | + |
| 39 | +### Exploratory Data Analysis |
| 40 | + |
| 41 | +- Aim is to understand the data |
| 42 | +- Descriptive statistics of central tendencies and variation |
| 43 | +- Basic plots of distributions / skewness (histograms) |
| 44 | +- Correlation plots |
| 45 | + |
| 46 | +### Feature Engineering / Transforming Variables |
| 47 | + |
| 48 | +- Reducing skew (log or other transformation) |
| 49 | +- Encoding category variables as dummies |
| 50 | +- Creating new predictor variables, interaction terms |
| 51 | +- Centering / Scaling Variables |
| 52 | + |
| 53 | +## Case Study: Predicting Rabies |
| 54 | + |
| 55 | +### Goal: |
| 56 | + |
| 57 | +Predict DALYs due to rabies in 'Asia' and 'Global' regions, using the `hmsidwR::rabies` dataset |
| 58 | + |
| 59 | +### Exploratory Data Analysis (EDA) |
| 60 | + |
| 61 | +- Dataset contains all cause and rabies mortality plus DALYs for the Asian and Global region, subdivided by year |
| 62 | + |
| 63 | +- Values have an estimate and upper and lower boundaries in separate columns |
| 64 | + |
| 65 | +- 240 observations across 7 variables. |
| 66 | + |
| 67 | +- Examining the data shows that death rates (`dx_rabies`) and DALYs (`dalys_rabies`) are different in magnitude and scale |
| 68 | + |
| 69 | +```{r rabiesdata, message=FALSE, warning=FALSE} |
| 70 | +
|
| 71 | +library(tidyverse) |
| 72 | +rabies <- hmsidwR::rabies %>% |
| 73 | + filter(year >= 1990 & year <= 2019) %>% |
| 74 | + select(-upper, -lower) %>% |
| 75 | + pivot_wider(names_from = measure, values_from = val) %>% |
| 76 | + filter(cause == "Rabies") %>% |
| 77 | + rename(dx_rabies = Deaths, dalys_rabies = DALYs) %>% |
| 78 | + select(-cause) |
| 79 | +
|
| 80 | +rabies %>% head() |
| 81 | +
|
| 82 | +``` |
| 83 | + |
| 84 | +- After scaling, these values are closer together in magnitude, avoiding the issue of larger variables dominating others in prediction |
| 85 | + |
| 86 | +```{r} |
| 87 | +
|
| 88 | +library(patchwork) |
| 89 | +
|
| 90 | +p1 <- rabies %>% |
| 91 | + ggplot(aes(x = year, group = location, linetype = location)) + |
| 92 | + geom_line(aes(y = dx_rabies), |
| 93 | + linewidth = 1) + |
| 94 | + geom_line(aes(y = dalys_rabies)) |
| 95 | +
|
| 96 | +p2 <- rabies %>% |
| 97 | + # apply a scale transformation to the numeric variables |
| 98 | + mutate(year = as.integer(year), |
| 99 | + across(where(is.double), scale)) %>% |
| 100 | + ggplot(aes(x = year, group = location, linetype = location)) + |
| 101 | + geom_line(aes(y = dx_rabies), |
| 102 | + linewidth = 1) + |
| 103 | + geom_line(aes(y = dalys_rabies)) |
| 104 | +
|
| 105 | +p1 + p2 |
| 106 | +
|
| 107 | +``` |
| 108 | + |
| 109 | +### Training and Resampling |
| 110 | + |
| 111 | +- The dataset was split into 80% training and 20% final test, stratified by location |
| 112 | +- The 80% training set was then used to create a series of 'folds' or resamples of the data |
| 113 | +- These folds can then be used to validate how well each model (and selected parameters) match unseen data |
| 114 | +- K-fold cross validation was used to generate 10 folds using the `vfold_cv()` function from the tidymodels package |
| 115 | + |
| 116 | +### Preprocessing |
| 117 | + |
| 118 | +- Handled using 'recipes' as part of tidymodels pipelines |
| 119 | +- **Recipe 0** - all predictors, no transformations [reference model] |
| 120 | +- **Recipe 1** - encoding of dummy variable for region, standardised numeric variables |
| 121 | +- **Recipe 2** - as recipe 2, with addition of method to reduce skewness of `dalys_rabies` outcome |
| 122 | +- Advantage of 'recipe' approach in tidymodels is that they can be piped / swapped out easily. |
| 123 | + |
| 124 | +### Multicollinearity |
| 125 | + |
| 126 | +- DALYs & mortality likely to be strongly correlated (DALYs = Years_life_lost + Years_lived_w_disability)) |
| 127 | +- All cause and specific cause mortality also will have some correlation |
| 128 | +- This can cause issues with some prediction methods, making it hard for the model to determine which variables have the best predictive power. |
| 129 | +- In this analysis, dealt with by the choice of prediction method: Random forests and GLM with lasso penalty both robust to multicollinearity |
| 130 | + |
| 131 | +### Model 1: Random forest |
| 132 | + |
| 133 | +- Specified using `rand_forest()` function within tidymodels framework |
| 134 | +- Hyperparameters tuned using cross-validation and `tune_grid()` / grid search |
| 135 | +- Optimal parameters gave RMSE 0.506 |
| 136 | +- Fig 7.4a shows close relationship between predictions and observed data |
| 137 | + |
| 138 | +[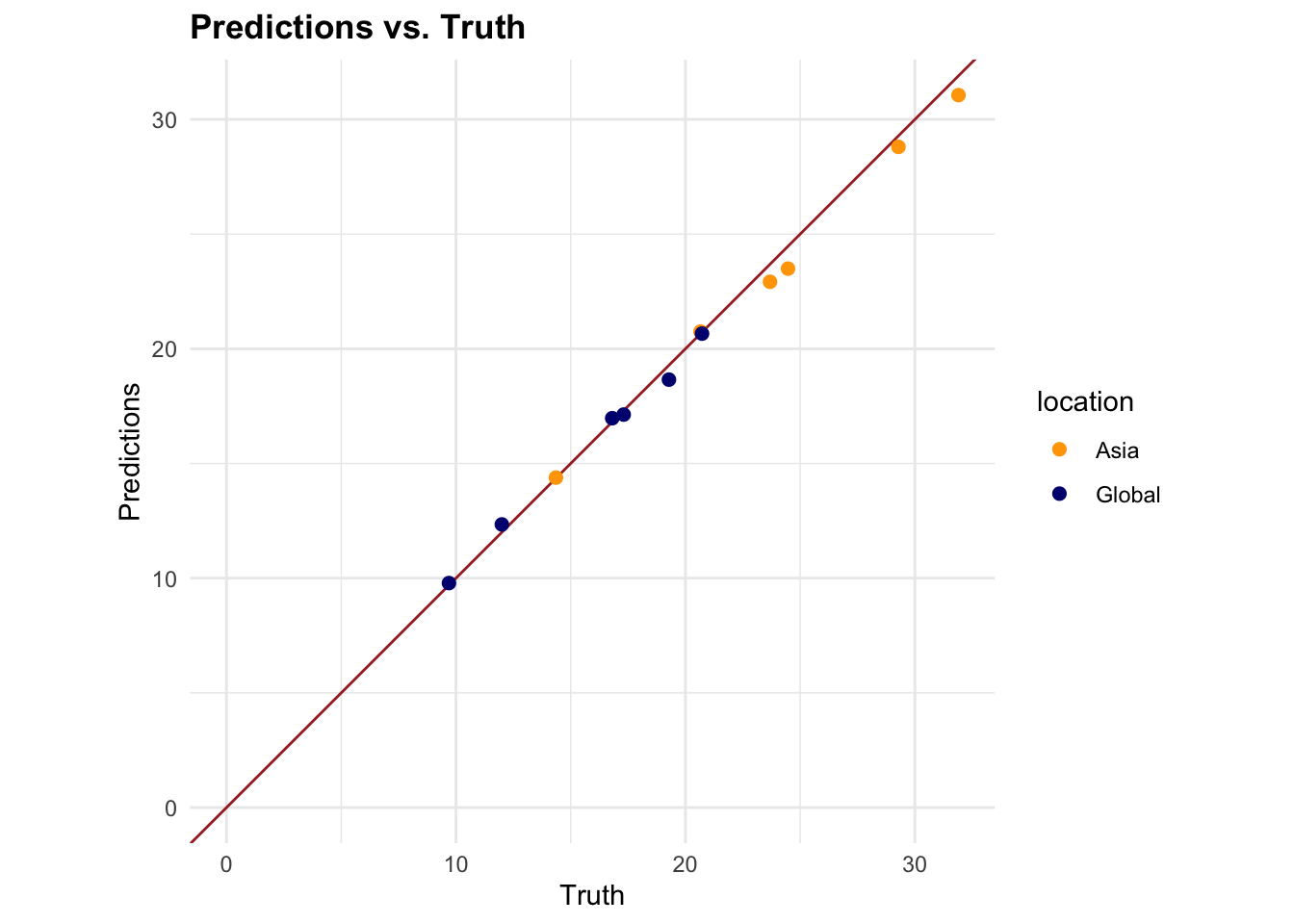](https://fgazzelloni.quarto.pub/06-techniques.html#fig-rf-predictions-1) |
| 139 | + |
| 140 | +### Model 2: GLM w lasso penalty |
| 141 | + |
| 142 | +- Generalised Linear Model with penalty term ($\lambda$) |
| 143 | +- Cross-validation process (as done for model 1) to tune $\lambda$ parameter |
| 144 | +- Results in lower RMSE than random forest |
| 145 | + |
| 146 | +### Additional models! |
| 147 | + |
| 148 | +- Last section showed code using `parsnip` package and `workflow_set()` to test more models |
| 149 | +- SVN with yeo_johnson transformation of output may actually improve on GLM (graded on RSME) |
| 150 | + |
| 151 | +## Summary |
| 152 | + |
| 153 | +This chapter focussed on ML techniques as a holistic analysis pipeline, not on individual ML algorithms or methods. Best practices are summarised at the end of the chapter: |
| 154 | + |
| 155 | +- Conduct exploratory data analysis to understand the underlying structure of the data and relationships between variables. |
| 156 | +- Apply feature engineering techniques to create new variables and enhance the model’s predictive power. |
| 157 | +- Select machine learning models that are contextually appropriate and robust for public health data analysis. Such as Random Forest, Generalised Linear Models, and others. |
| 158 | +- Use parameter calibration techniques such as cross-validation, regularisation, monte carlo, and grid search to optimise model performance. |
| 159 | +- Evaluate model performance using appropriate metrics and visualisation tools to assess predictive accuracy and relevance. |
| 160 | + |
| 161 | +TLDR: It's not just about applying the individual ML model but about considering the goals, dataset, preprocessing, calibration and evaluation of the model. |
| 162 | + |
| 163 | +## Meeting Videos {.unnumbered} |
| 164 | + |
| 165 | +### Cohort 1 {.unnumbered} |
15 | 166 |
|
16 | 167 | `r knitr::include_url("https://www.youtube.com/embed/URL")` |
17 | 168 |
|
18 | 169 | <details> |
19 | | -<summary> Meeting chat log </summary> |
20 | 170 |
|
21 | | -``` |
| 171 | +<summary>Meeting chat log</summary> |
| 172 | + |
| 173 | +``` |
22 | 174 | LOG |
23 | 175 | ``` |
| 176 | + |
24 | 177 | </details> |
0 commit comments