As a data scientist, we all know that unglamorous data manipulation is 90% of the work. Two of the most common data manipulation tools are SQL and Pandas. In this blog, we'll compare the performance of pandas and sqlite, a simple form a SQL favored by data scientists.
Let's find out the tasks at which each of these excel. Below, we compare
Python's pandas to sqlite for some common data analysis operations: sort,
select, load, join, filter, and group by.
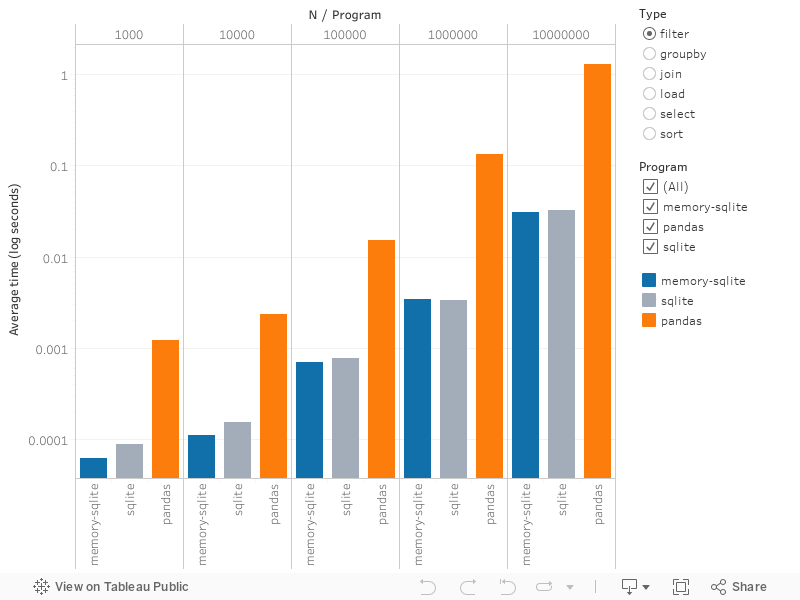
Note that the axis is logarithmic, so that raw differences are more pronounced.
<script type='text/javascript'> var divElement = document.getElementById('viz1487191497220'); var vizElement = divElement.getElementsByTagName('object')[0]; vizElement.style.width='100%';vizElement.style.height=(divElement.offsetWidth*0.75)+'px'; var scriptElement = document.createElement('script'); scriptElement.src = 'https://public.tableau.com/javascripts/api/viz_v1.js'; vizElement.parentNode.insertBefore(scriptElement, vizElement); </script>For the analysis, we ran the six tasks 10 times each, for 5 different sample
sizes, for each of 3 programs: pandas, sqlite, and memory-sqlite (where
database is in memory instead of on disk). See below for the definitions of
each task.
Our sample data was randomly generated. Here's what it looks like:
sql_vs_pandas$ head -n 5 data/sample.100.csv
qqFjQHQc,c,1981,82405.59262172286
vILuhVGz,a,1908,27712.27152250119
mwCjpoOF,f,1992,58974.38538762843
kGbriYAK,d,1927,42258.24179716961
MeoxuJng,c,1955,96907.56416314292
This consists of a random string of 8 characters, a random single character (for the filtering operation), a random integer simulating a year (1900-2000), and a uniform random float value between 10000 and 100000.
sqlite or memory-sqlite is faster for the following tasks:
selecttwo columns from data (<.1 millisecond for any data size forsqlite.pandasscales with the data, up to just under 0.5 seconds for 10 million records)filterdata (>10x-50x faster withsqlite. The difference is more pronounced as data grows in size)sortby single column:pandasis always a bit slower, but this was the closest
pandas is faster for the following tasks:
groupbycomputation of a mean and sum (significantly better for large data, only 2x faster for <10k records)loaddata from disk (5x faster for >10k records, even better for smaller data)joindata (2-5x faster, but slower for smallest data set of 1000 rows)
Comparing memory-sqlite vs. sqlite, there was no meaningful difference,
especially as data size increased.
There is no significant speedup from loading sqlite in its own shell vs. via
pandas.
Overall, joining and loading data is the slowest whereas select and filter are
generally the fastest. Further, pandas seems to be optimized for group-by
operations, where it performs really well (group-by is pandas' second fastest
operation for larger data).
Note that this analysis assumes you are equally proficient in writing code with both! But these results could encourage you to learn the tool that you are less familiar with, if the performance gains are significant.
All code is on our GitHub page.
Below are the definitions of our six tasks: sort, select, load, join, filter,
and group by (see driver/sqlite_driver.py or driver/pandas_driver.py).
sqlite is first, followed by pandas:
def sort(self):
self._cursor.execute('SELECT * FROM employee ORDER BY name ASC;')
self._conn.commit()
def sort(self):
self.df_employee.sort_values(by='name')
def select(self):
self._cursor.execute('SELECT name, dept FROM employee;')
self._conn.commit()
def select(self):
self.df_employee[["name", "dept"]]
def load(self):
self._cursor.execute('CREATE TABLE employee (name varchar(255), dept char(1), birth int, salary double);')
df = pd.read_csv(self.employee_file)
df.columns = employee_columns
df.to_sql('employee', self._conn, if_exists='replace')
self._cursor.execute('CREATE TABLE bonus (name varchar(255), bonus double);')
df_bonus = pd.read_csv(self.bonus_file)
df_bonus.columns = bonus_columns
df_bonus.to_sql('bonus', self._conn, if_exists='replace')
def load(self):
self.df_employee = pd.read_csv(self.employee_file)
self.df_employee.columns = employee_columns
self.df_bonus = pd.read_csv(self.bonus_file)
self.df_bonus.columns = bonus_columns
def join(self):
self._cursor.execute('SELECT employee.name, employee.salary + bonus.bonus '
'FROM employee INNER JOIN bonus ON employee.name = bonus.name')
self._conn.commit()
def join(self):
joined = self.df_employee.merge(self.df_bonus, on='name')
joined['total'] = joined['bonus'] + joined['salary']
def filter(self):
self._cursor.execute('SELECT * FROM employee WHERE dept = "a";')
self._conn.commit()
def filter(self):
self.df_employee[self.df_employee['dept'] == 'a']
def groupby(self):
self._cursor.execute('SELECT avg(birth), sum(salary) FROM employee GROUP BY dept;')
self._conn.commit()
def groupby(self):
self.df_employee.groupby("dept").agg({'birth': np.mean, 'salary': np.sum})
We used pandas version 0.19.1 and sqlite version 3.13.0. All tests
were ran on Digital Ocean Ubuntu 14.04 with 16GB memory and 8 core CPU.
For resource on becoming a data scientist, check out our blog, particular this article on preparing for our free data science fellowship and this one on data manipulating data like a professional data scientist. And if you're looking for a class, consider our convenient after-work online instructor lead data science foundations course geared towards working professionals or our free data science fellowship for people with advanced degrees.
To learn more how sqlite works, check
out
this awesome blog series.
Here is
a
syntax comparison between
pandas and sql.