One can use the Colab to evaluate our latest models.
Model zoom: https://zenodo.org/record/8031783
📘Documentation | 🛠️Installation | 🚀Model Zoo | 🆕News
This repository is an open-source project for benchmarking structure-based protein design methods, which provides a variety of collated datasets, reprouduced methods, novel evaluation metrics, and fine-tuned models that are all integrated into one unified framework. It also contains the implementation code for the paper:
ProteinInvBench: Benchmarking Protein Design on Diverse Tasks, Models, and Metrics
Zhangyang Gao, Cheng Tan, Yijie Zhang, Xingran Chen, Stan Z. Li.
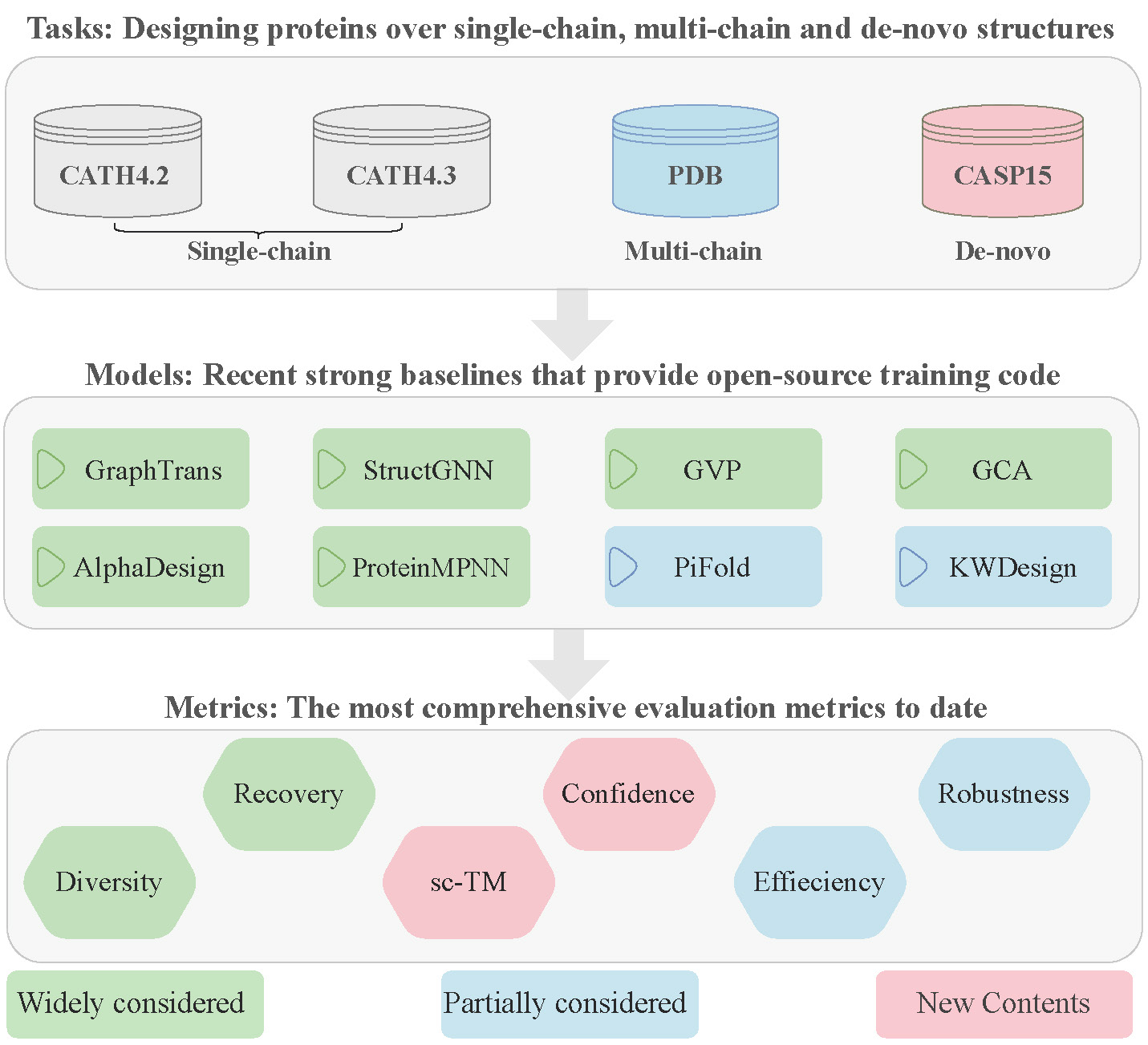
ProteinInvBench is the first comprehensive benchmark for protein design. The main contributions of our paper could be listed as four points below:
- Tasks: We extend recent impressive models from single-chain protein design to the scenarios of multi-chain and de-novoprotein design.
- Models: We integrate recent impressive models into a unified framework for efficiently reproducing and extending them to custom tasks.
- Metrics: We incorporate new metrics such as confidence, sc-TM, and diversity for protein design, and integrate metrics including recovery, robustness, and efficiency to formulate a comprehensive evaluation system.
- Benchmark: We establish the first comprehensive benchmark of protein design, providing insights into the strengths and weaknesses of different methods.
pip install pyg_lib torch_scatter torch_sparse torch_cluster torch_spline_conv -f https://data.pyg.org/whl/torch-2.4.0+cu121.html
pip install -r requirements.txt
pip install PInvBench==0.1.0Major Features
- Unified Code Framework
ProteinInvBench integrates the protein design system pipeline into a unified framework. From data preprocessing to model training, evaluating and result recording, all the methods collected in this paper could be conducted in the same way, which simplifies further analysis to the problem. In detail, ProteinInvBench decomposes computational protein design algorithms into
methods(training and prediction),models(network architectures), andmodules.Users can develop their own algorithms with flexible training strategies and networks for different protein design tasks. - Comprehensive Model Implementation ProteinInvBench collects a wide range of recent impressive models together with the datasets and reproduces all the methods in each of the datasets with restricted manners.
- Standard Benchmarks ProteinInvBench supports standard benchmarks of computational protein design algorithms with various evaluation metrics, including novel ones such as confidence, diversity, and sc-TM. The wide range of evaluataion metrics helps to have a more comprehensive understanding of different protein design algorithms.
Code Structures
run/contains the experiment runner and dataset configurations.configs/contains the model configurationsopencpd/corecontains core training plugins and metrics.opencpd/datasetscontains datasets and dataloaders.opencpd/methods/contains collected models for various protein design methods.opencpd/models/contains the main network architectures of various protein design methods.opencpd/modules/contains network modules and layers.opencpd/utils/contains some details in each model.tools/contains the executable python files and script files to prepare the dateset and save checkpoints to the model.
[2023-06-16] ProteinInvBench v0.1.0 is released.
This project has provided an environment setting file of conda, users can easily reproduce the environment by the following commands:
git clone https://github.com/A4Bio/OpenCPD.git
cd opencpd
conda env create -f environment.yml
conda activate opencpd
python setup.py developObtaining Dataset
The processed datasets could be found in the releases.
To note that, due to the large file size, ProteinMPNN dataset was uploaded in a separate file named mpnn.tar.gz, others could be found in data.tar.gz
Model Training
python main.py --method {method} We support various protein design methods and will provide benchmarks on various protein datasets. We are working on adding new methods and collecting experiment results.
-
Protein Design Methods.
Currently supported methods
The models and their corresponding results and checkpoints can be downloaded here
- GraphTrans (NeurIPS'2021)
- StructGNN (NeurIPS'2020)
- GVP
- GCA (ICASSP'2023)
- AlphaDesign
- ProteinMPNN (Science)
- PiFold (ICLR'2023)
- KWDesign
Currently supported datasets
To download the processed datasets, please click here The details of the datasets could be found in dataset.md
- CATH
- PDB
- CASP15
Currently supported evaluation metrics
- Recovery
- Confidence
- Diversity
- sc-TM
- Robustness
- Efficiency
This project is released under the Apache 2.0 license. See LICENSE for more information.
ProteinInvBench is an open-source project for structure-based protein design methods created by researchers in CAIRI AI Lab. We encourage researchers interested in protein design and other related fields to contribute to this project!
@inproceedings{
gao2023proteininvbench,
title={ProteinInvBench: Benchmarking Protein Inverse Folding on Diverse Tasks, Models, and Metrics},
author={Zhangyang Gao and Cheng Tan and Yijie Zhang and Xingran Chen and Lirong Wu and Stan Z. Li},
booktitle={Thirty-seventh Conference on Neural Information Processing Systems Datasets and Benchmarks Track},
year={2023},
url={https://openreview.net/forum?id=bqXduvuW5E}
}
For adding new features, looking for helps, or reporting bugs associated with ProteinInvBench, please open a GitHub issue and pull request with the tag "new features", "help wanted", or "enhancement". Feel free to contact us through email if you have any questions.
- Zhangyang Gao ([email protected]), Westlake University & Zhejiang University
- Cheng Tan ([email protected]), Westlake University & Zhejiang University
- Yijie Zhang ([email protected]), McGill University
- Xingran Chen([email protected]), University of Michigan
- Switch code to torch_lightning framework (Done)
- Deploy code to public server
- Support pip installation
export PYTHONPATH=path/ProteinInvBench
python PInvBench/evaluation_tools/InverseFolding.py --pdb_path test_pdbs --sv_fasta_path test.fasta --model UniIF --topk 5 --temp 1.0For each pdb file in test_pdbs, we use UniIF to design the corresponding top-k sequence and save the results in test.fasta. The sampling tempertature is 1.0.