Important
👉 Now part of Docling!
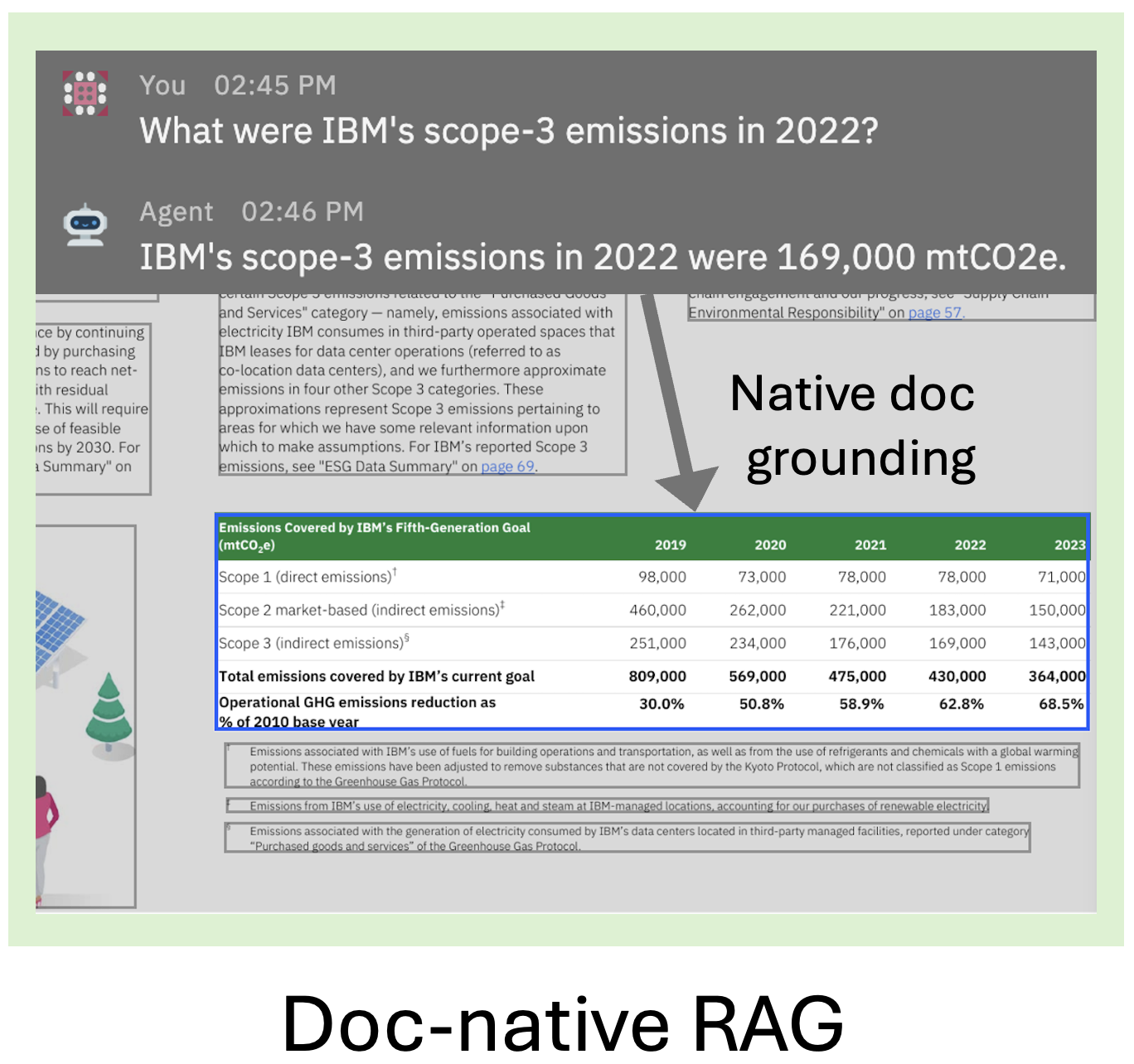
Easily build document-native generative AI applications, such as RAG, leveraging Docling's efficient PDF extraction and rich data model — while still using your favorite framework, 🦙 LlamaIndex or 🦜🔗 LangChain.
- 🧠 Enables rich gen AI applications by providing capabilities on native document level — not just plain text / Markdown!
- ⚡️ Leverages Docling's conversion quality and speed.
- ⚙️ Plug-and-play integration with LlamaIndex and LangChain for building powerful applications like RAG.
To use Quackling, simply install quackling from your package manager, e.g. pip:
pip install quacklingQuackling offers core capabilities (quackling.core), as well as framework integration components (quackling.llama_index and quackling.langchain). Below you find examples of both.
Here is a basic RAG pipeline using LlamaIndex:
Note
To use as is, first pip install llama-index-embeddings-huggingface llama-index-llms-huggingface-api
additionally to quackling to install the models.
Otherwise, you can set EMBED_MODEL & LLM as desired, e.g. using
local models.
import os
from llama_index.core import VectorStoreIndex
from llama_index.embeddings.huggingface import HuggingFaceEmbedding
from llama_index.llms.huggingface_api import HuggingFaceInferenceAPI
from quackling.llama_index.node_parsers import HierarchicalJSONNodeParser
from quackling.llama_index.readers import DoclingPDFReader
DOCS = ["https://arxiv.org/pdf/2206.01062"]
QUESTION = "How many pages were human annotated?"
EMBED_MODEL = HuggingFaceEmbedding(model_name="BAAI/bge-small-en-v1.5")
LLM = HuggingFaceInferenceAPI(
token=os.getenv("HF_TOKEN"),
model_name="mistralai/Mistral-7B-Instruct-v0.3",
)
index = VectorStoreIndex.from_documents(
documents=DoclingPDFReader(parse_type=DoclingPDFReader.ParseType.JSON).load_data(DOCS),
embed_model=EMBED_MODEL,
transformations=[HierarchicalJSONNodeParser()],
)
query_engine = index.as_query_engine(llm=LLM)
result = query_engine.query(QUESTION)
print(result.response)
# > 80K pages were human annotatedYou can also use Quackling as a standalone with any pipeline. For instance, to split the document to chunks based on document structure and returning pointers to Docling document's nodes:
from docling.document_converter import DocumentConverter
from quackling.core.chunkers import HierarchicalChunker
doc = DocumentConverter().convert_single("https://arxiv.org/pdf/2408.09869").output
chunks = list(HierarchicalChunker().chunk(doc))
# > [
# > ChunkWithMetadata(
# > path='$.main-text[4]',
# > text='Docling Technical Report\n[...]',
# > page=1,
# > bbox=[117.56, 439.85, 494.07, 482.42]
# > ),
# > [...]
# > ]- Milvus basic RAG (dense embeddings)
- Milvus hybrid RAG (dense & sparse embeddings combined e.g. via RRF) & reranker model usage
- Milvus RAG also fetching native document metadata for search results
- Local node transformations (e.g. embeddings)
- ...
Please read Contributing to Quackling for details.
If you use Quackling in your projects, please consider citing the following:
@techreport{Docling,
author = "Deep Search Team",
month = 8,
title = "Docling Technical Report",
url = "https://arxiv.org/abs/2408.09869",
eprint = "2408.09869",
doi = "10.48550/arXiv.2408.09869",
version = "1.0.0",
year = 2024
}The Quackling codebase is under MIT license. For individual component usage, please refer to the component licenses found in the original packages.