NOTE: This project is no longer supported. Consider using http://genepio.org/DataHarmonizer/main.html instead
The INCA table editor enables biomedical curators to create new ontology term requests compositionally from a configurable set of existing ontologies and terminologies and within a familiar spreadsheet-like interface. Because directly editing ontology owl files requires considerable training (not to mention patience and QC), spreadsheet-based methods for term requests are not new; however, what INCA Table Editor provides a combination of three key innovations: configurable autocomplete, design-pattern driven logic, and (planned) GitHub integration. This democratizes and accelerates the term request process while also avoiding errors of logic right at the source. Each new term request is complete and logically valid, and is issued a generic IRI until such a time as it can be given an actual namespace and ID in an appropriate ontology.
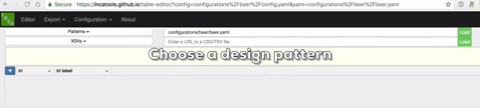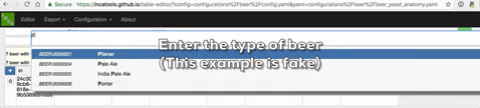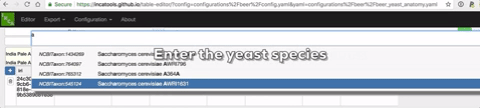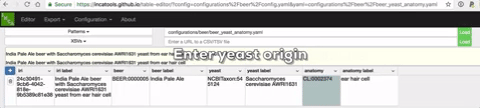
The formal pattern-based approach we use is the Dead Simple Ontology Design Patterns (DOSDP) published here (PMC5460348). The patterns are written in YAML. An example is here.
Using a different INCA configuration, the DOSDP can also be leveraged for instance-level annotations, such as between diseases and phenotypes, or genes and phenotypes, or a patient and phenotypes. This kind of annotation use case is sometimes called "A-box"; where as the terminology use case above is sometimes called "T-box".
A live demo of the site can be seen at https://incatools.github.io/table-editor/; try out the beer example yourself (for other design patterns you will need to deploy it per instructions below). We welcome feedback at https://github.com/INCATools/table-editor/issues.
This lightweight web application is intended to be deployed as a static single-page website, where the site can then be used to view and edit spreadsheet-formatted data while supporting autocomplete and selection from semantically associated ontologies and dictionaries.
Both TSV and CSV are supported, and are collectively referred to as "XSV" in the software and documentation.
The initial specification for this table-editor is drawn from INCATools/intelligent-concept-assistant#2
- Autocomplete is hardcoded to use the Monarch Initiative autocomplete server, rather than being dynamically parameterized via a URL parameter.
- Selecting a cell should pop up the autocomplete menu immediately, rather than requiring a character be typed.
- Currently, the suffix ' label' on a column indicates that it is non-editable and derived from the root column name (e.g., 'location label' is derived from 'location'). This heuristic will be replaced by design guidance provided in the DOSDP YAML file.
- Some of the buttons and panels in the UI are to help debug this application, and not necessarily part of the final UI. For example, the display of the raw and parsed YAML is unnecessary.
The app has a few preloaded examples of YAML and CSV.
A local XSV file can be viewed through the app by using the 'Load File' button or by dragging and dropping a file onto the dropzone
A remote XSV file can be loaded via URL.
Note that if the remote file is hosted on a website that does not support CORS, then the request will be rejected. Such files can be copied locally and then viewed that way, however.
The table-editor app is designed so that the URL pointing to the app can be amended with an optional url parameter that points to a XSV file, subject to the same restrictions as Load URL above. For example, suppose this app is hosted on https://github.com/INCATools/table-editor and a desired Table file is hosted on http://www.example.com/MyTableFile.gv. Then the following URL will launch the table-editor app and cause it to load and render the specified file:
https://github.com/INCATools/table-editor?url=http://www.example.com/MyTableFile.gv
- Now that the prototype user interface and application behavior have been established, we need to simplify the current workflow which requires a user to manually manage the reading and writing of their curated TSV/CSV ontology files. We will do this by implementing a GitHub integration that enables the CSV/TSV to be imported directly from a GitHub branch and then exported back to that branch as a new revision. We will not be creating a GitHub 'pull request', as we have discovered that some of our users do not use the 'pull request' mechanism during their GitHub usage. However, all of our users that use GitHub would be able to use the simpler 'read from branch' and 'write to branch' capability.
- Better documentation about the purpose and usage of IncaForm and tetool.
- Use browser local storage to reduce the risk that a user would lose their 'work session' with an accidental premature browser refresh.
- Incorporate recent/developing changes to DOSDP to support annotation-mode curation. Currently, annotation-mode curation via IncaForm is specified with an IncaForm-specific configuration file format. It is desirable to instead use DOSDP for both T-box ontology curation, as well as for A-box curation and annotation.
- Use IncaForm (in annotation mode) to support annotation work for Fanconi anemia variants and models in the literature.
This is what I use, you may get lucky with slightly older/newer versions. Don't even bother trying Node 0.1x.
- NodeJS 4.5.0
- NPM 3.10.8
Tested on MacOSX Safari, Chrome and FireFox. Requires some form of http-server. npm run dev will invoke the WebPack server for auto-bundling during development, and this is sufficient for demo purposes.
cd table-editor/ # If you aren't alread there
npm install
npm run build # 'npm run fastbuild' to avoid minification
npm run dev
open http://localhost:8085/webpack-dev-server/table-editor # On MacOSX
# Alternatively, point your browser to:
# http://localhost:8085/webpack-dev-server/table-editor
#
- Planteome's Plant Experimental Conditions Ontology: https://github.com/Planteome/plant-experimental-conditions-ontology
- Planteome's Plant Trait Ontology: https://github.com/Planteome/plant-trait-ontology
- Planteome's Ontology of Plant Stress: https://github.com/Planteome/ontology-of-plant-stress
- Environment Ontology, Exposures: https://github.com/EnvironmentOntology/environmental-exposure-ontology
- Angular
- Bootstrap
- https://github.com/nodeca/js-yaml
- https://github.com/danialfarid/ng-file-upload
- https://github.com/mholt/PapaParse
- https://github.com/angular-ui/ui-grid
- This project is supported by NIH:U01-HG009453-01: An Intelligent Concept Agent for Assisting with the Application of Metadata