 CytoDataFrame extends Pandas functionality to help display single-cell profile data alongside related images.
CytoDataFrame extends Pandas functionality to help display single-cell profile data alongside related images.
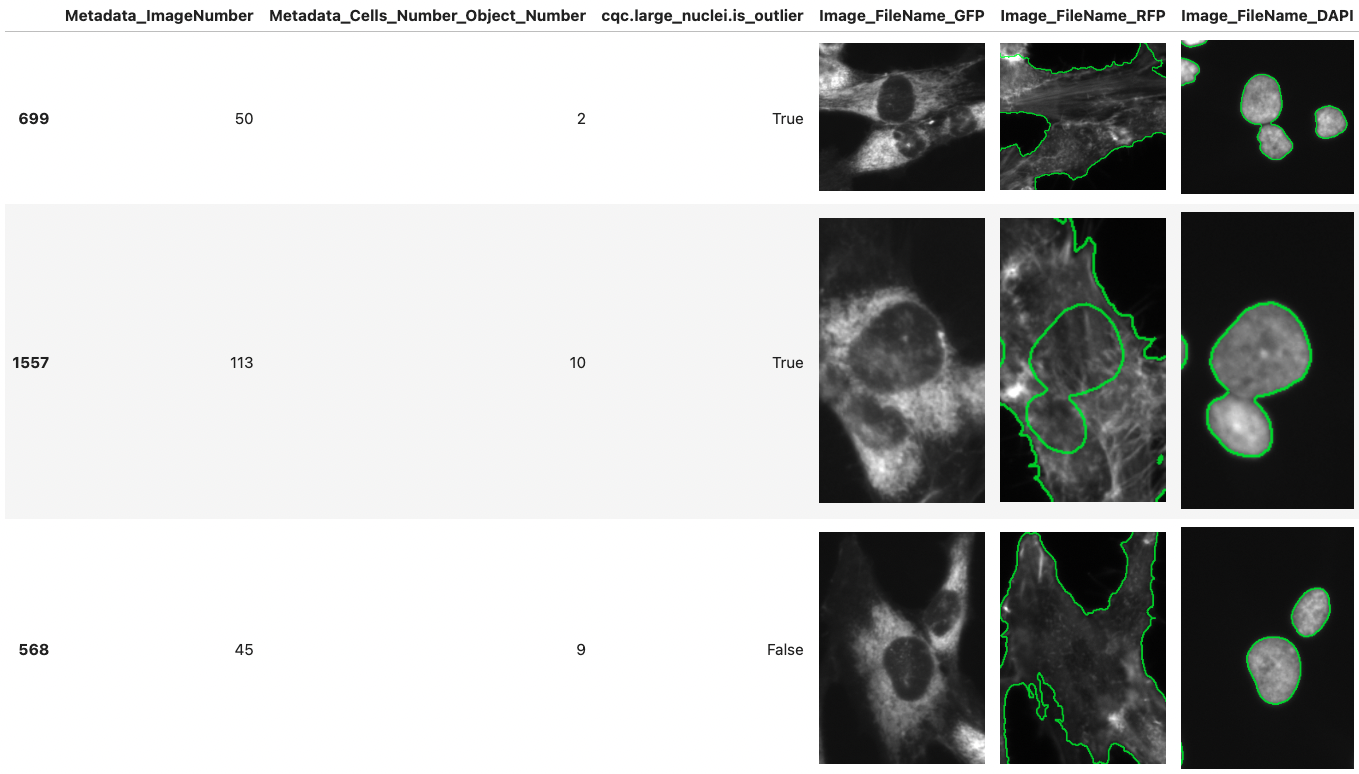
CytoDataFrame is an advanced in-memory data analysis format designed for single-cell profiling, integrating not only the data profiles but also their corresponding microscopy images and segmentation masks. Traditional single-cell profiling often excludes the associated images from analysis, limiting the scope of research. CytoDataFrame bridges this gap, offering a purpose-built solution for comprehensive analysis that incorporates both the data and images, empowering more detailed and visual insights in single-cell research.
CytoDataFrame development began within coSMicQC - a single-cell profile quality control package.
Install CytoDataFrame from source using the following:
# install from pypi
pip install cytodataframe
# or install directly from source
pip install git+https://github.com/WayScience/CytoDataFrame.gitPlease see our contributing documentation for more details on contributions, development, and testing.



