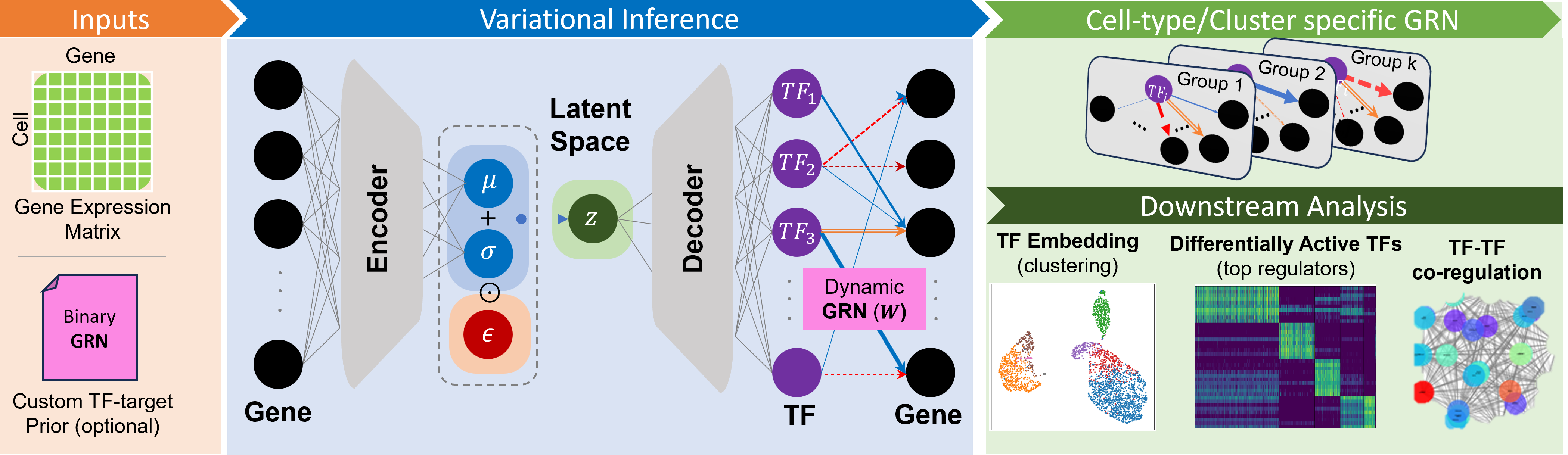
Single-Cell Regulatory-Embedded Variational Inference of Transcription Factor Activity from Gene Expression
scRegulate is a powerful tool designed for the inference of transcription factor activity from single cell/nucleus RNA data using advanced generative modeling techniques. It leverages a unified learning framework to optimize the modeling of cellular regulatory networks, providing researchers with accurate insights into transcriptional regulation. With its efficient clustering capabilities, scRegulate facilitates the analysis of complex biological data, making it an essential resource for studies in genomics and molecular biology.
For further information and example tutorials, please check our documentation:
If you have any questions or concerns, feel free to open an issue.
scRegulate is implemented in the PyTorch framework. Running scRegulate on CUDA is highly recommended if available.
Before installing and running scRegulate, ensure you have the following libraries installed:
- PyTorch (version 2.0 or higher)
Install with the exact command from the PyTorch “Get Started” page for your OS, Python version and (optionally) CUDA toolkit. - NumPy (version 1.23 or higher)
- Scanpy (version 1.9 or higher)
- Anndata (version 0.8 or higher)
You can install these dependencies using pip:
pip install torch numpy scanpy anndataOption 1:
You can install scRegulate via pip for a lightweight installation:
pip install scregulateOption 2:
Alternatively, if you want the latest, unreleased version, you can install it directly from the source on GitHub:
pip install git+https://github.com/YDaiLab/scRegulate.gitOption 3:
For users who prefer Conda or Mamba for environment management, you can install scRegulate along with extra dependencies:
Conda:
conda install -c zandigohar scregulateMamba:
mamba create -n scRegulate -c zandigohar scregulateQ1: Do I need a GPU to run scRegulate?
No, a GPU is not required. However, using a CUDA-enabled GPU is strongly recommended for faster training and inference, especially with large datasets.
Q2: How do I know if I can use a GPU with scRegulate?
There are two quick checks:
-
System check
In your terminal, runnvidia-smi. If you see your GPU listed (model, memory, driver version), your machine has an NVIDIA GPU with the driver installed. -
Python check
In a Python shell, run:import torch print(torch.cuda.is_available()) # True means PyTorch can see your GPU print(torch.cuda.device_count()) # How many GPUs are usable
Q3: Can I use scRegulate with Seurat or R-based tools?
scRegulate is written in Python and works directly with AnnData objects (e.g., from Scanpy). You can convert Seurat objects to AnnData using tools like SeuratDisk.
Q4: How can I visualize inferred TF activities?
TF activities inferred by scRegulate are stored in the obsm slot of the AnnData object. You can use scanpy.pl.embedding, scanpy.pl.heatmap, or export the matrix for custom plots.
Q5: What kind of prior networks does scRegulate accept?
scRegulate supports user-provided gene regulatory networks (GRNs) in CSV or matrix format. These can be curated from public databases or inferred from ATAC-seq or motif analysis.
Q6: Can I use scRegulate for multi-omics integration?
Not directly. While scRegulate focuses on TF activity from RNA, you can incorporate priors derived from other omics (e.g., ATAC) to guide the model.
Q7: What file formats are supported?
scRegulate works with .h5ad files (AnnData format). Input files should contain gene expression matrices with proper normalization.
Q8: How do I cite scRegulate?
See the Citation section below for the latest reference and preprint link.
Q9: How can I reproduce the paper’s results?
See our Reproducibility Guide for step-by-step instructions. Then run scregulate.
scRegulate manuscript is currently under peer review.
If you use scRegulate in your research, please cite:
Mehrdad Zandigohar, Jalees Rehman and Yang Dai (2025). scRegulate: Single-Cell Regulatory-Embedded Variational Inference of Transcription Factor Activity from Gene Expression, Bioinformatics Journal (under review).
📄 Read the preprint on bioRxiv: 10.1101/2025.04.17.649372
scRegulate was developed and is actively maintained by Mehrdad Zandigohar as part of his PhD research at the University of Illinois Chicago (UIC), in the lab of Dr. Yang Dai.
📬 For private questions, please email: [email protected]
🤝 For collaboration inquiries, please contact PI: Dr. Yang Dai ([email protected])
Contributions, feature suggestions, and feedback are always welcome!
The code in scRegulate is licensed under the MIT License, which permits academic and commercial use, modification, and distribution.
Please note that any third-party dependencies bundled with scRegulate may have their own respective licenses.