*Named after Frank W. Preston (1896-1989) and the Prestonian shortfall, one of the "Seven Shortfalls that Beset Large-Scale Knowledge of Biodiversity" as described by Hortal et al. 2015.
When using Preston, please cite:
Elliott M.J., Poelen J.H., Fortes J.A.B. (2020). Toward Reliable Biodiversity Dataset References. Ecological Informatics. https://doi.org/10.1016/j.ecoinf.2020.101132 hash://sha256/136c3c1808bcf463bb04b11622bb2e7b5fba28f5be1fc258c5ea55b3b84f482c
and/or
Elliott M.J., Poelen, J.H. & Fortes, J.A.B. (2023) Signing data citations enables data verification and citation persistence. Sci Data. https://doi.org/10.1038/s41597-023-02230-y hash://sha256/f849c870565f608899f183ca261365dce9c9f1c5441b1c779e0db49df9c2a19d
toc / quickstart / introduction / usage / install / use cases / architecture / funding / (data) publications
Use homebrew to install Preston on your Mac via:
brew install globalbioticinteractions/globi/prestonTo install Preston on Debian, Ubuntu, etc, you can use apt via:
sudo apt update
sudo apt upgrade
curl -L https://github.com/bio-guoda/preston/releases/latest/download/preston.deb > preston.deb
sudo apt install ./preston.debTo remove type sudo apt remove preston.
To install Preston on RedHat, Fedora, etc, dnf can be used:
curl -L https://github.com/bio-guoda/preston/releases/latest/download/preston.rpm > preston.rpm
sudo dnf install ./preston.rpmTo remove type sudo dnf remove preston.
To install a preston release on your linux/mac:
sudo sh -c '(curl -L https://github.com/bio-guoda/preston/releases/latest/download/preston.jar) > /usr/local/bin/preston && chmod +x /usr/local/bin/preston && preston config-manpage' && preston versionNote that a homebrew formula (Mac), preston.deb (Debian, Ubuntu, etc) and preston.rpm (RedHat, Fedora, etc) packages are also available.
Then, visit jhpoelen.nl/bees or github.com/bio-guoda/preston-amazon for worked out examples.
Alternatively, run:
cd [some_dir]
preston clone "https://jhpoelen.nl/bees/data"
preston cat hash://sha256/edde5b2b45961e356f27b81a3aa51584de4761ad9fa678c4b9fa3230808ea356 > bee.jpg
and open bee.jpg to view your local copy of the following headshot of Nomadopsis puellae specimen MCZ:Ent:17219 from the Museum of Comparative Zoology, Harvard University CC-BY-NC-SA:
You can find more (exact) copies of this image via hash-archive.
Preston is an open-source software system that captures and catalogs biodiversity datasets. It enables reproducible research: scientists can use Preston to work with a uniquely identifiable, versioned copy of all or parts of GBIF-indexed datasets; dataset registry lookups: institutions can use Preston to check if and when their collections have been indexed and made available through iDigBio; cross-network analysis: biodiversity informatics researchers can use Preston to evaluate dataset overlap between GBIF and iDigBio; and finally, decentralized dataset archival: archivists can distribute Preston-generated biodiversity dataset archives across the world.
Preston uses the PROV and PAV ontologies to model the actors, activities and entities involved in discovery, access and change control of digital biodiversity datasets. In addition, Preston uses content-addressed storage and SHA256 hashes to uniquely identify and store content. A hexastore-like index is used to navigate a local graph of biodiversity datasets. Preston is designed to work offline and can be cloned, copied and moved across storage media with existing tools and infrastructures like rsync, dropbox, the internet archive or thumbdrives. In addition to versioned copies of uniquely identifiable original ABCD-A, DWC-A and EML files, Preston also keeps track of the GBIF, iDigBio and BioCASe registries to help retain the relationships between the institutions to keep a detailed record of provenance.
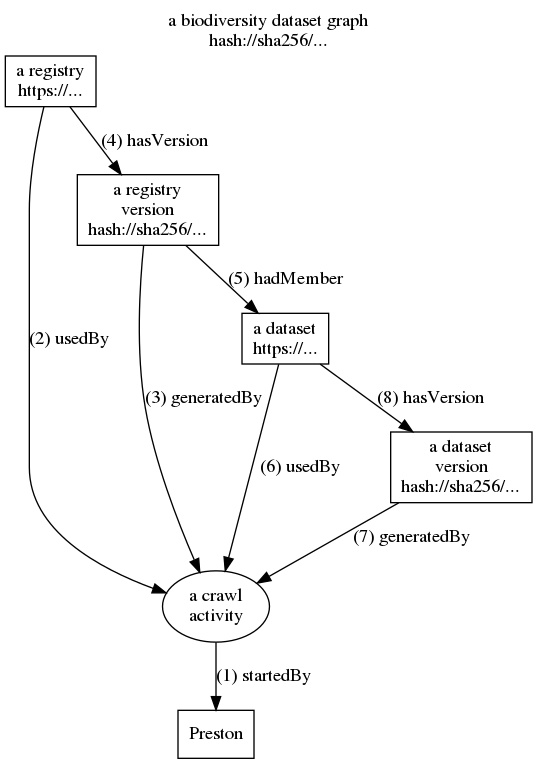
To capture and catalog biodiversity datasets, Preston performs a crawl process (see diagram below) for each institution which stores biodiversity datasets:
- A crawl activity is started (1)
- The registry (set of datasets) of an institution is requested and downloaded (2)
- The sha-256 hash is computed for the downloaded registry (3) (4)
- A list of the registry's datasets is created and each dataset is related to the registry (5)
- For each dataset in the registry:
- The information about the dataset is downloaded (6)
- For the downloaded dataset, it is computed the sha-256 hash (7) (8)
- The crawl activity finishes: log is completed.
The process diagram below shows how Preston starts crawls to download copies of biodiversity registries and their datasets. A detailed log of the crawl activities is recorded to describe what data was discovered and how. This activity log is referred to as the history of a biodiversity dataset graph. The numbers indicate the sequence of events. Click on the image to enlarge.
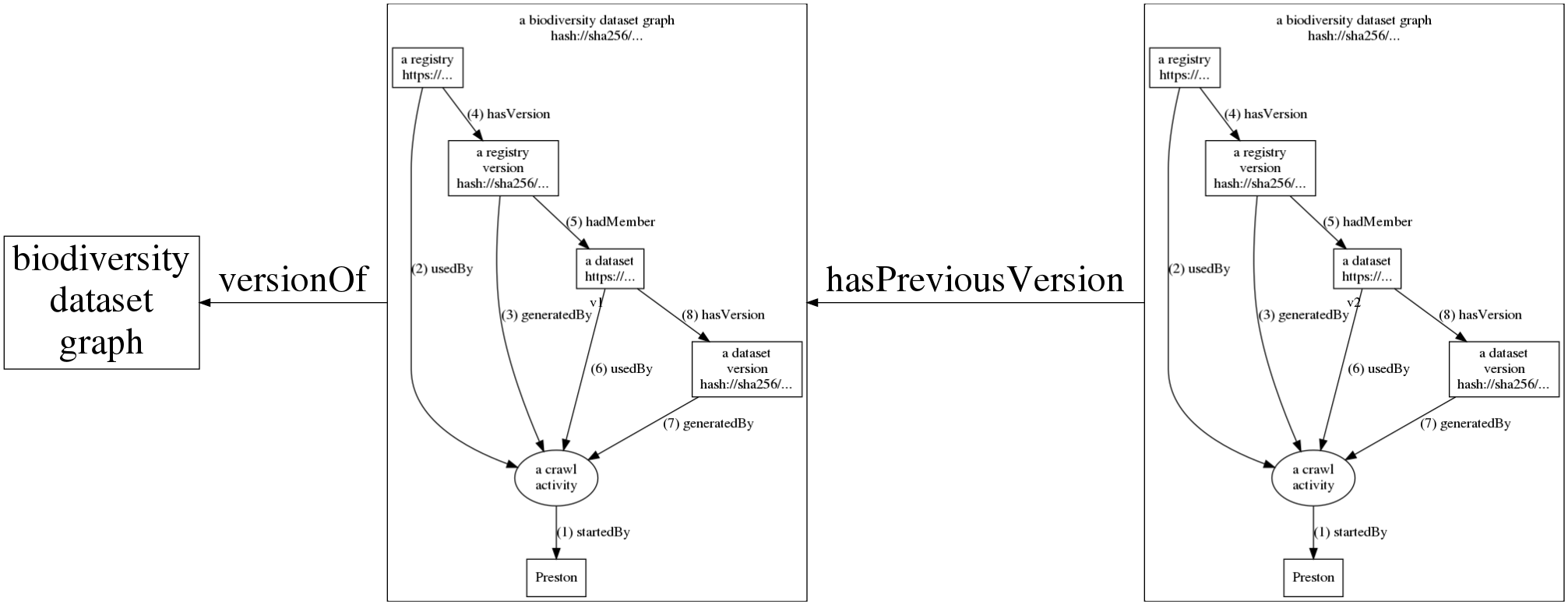
The figure above shows how Preston starts (1) a crawl activity. This crawl activity then accesses (2) a registry to save (3,4) a snapshot (or version) of it. Now, datasets referenced in this registry version are accessed, downloaded and saved (6,7,8). After all this, the crawl activity saves the log that contains its activities (1-8) as a version of a biodiversity dataset and linked to previous versions (see figure below). This log can be used to retrace the steps of the crawl activity to reconstruct the relationships between the registries, datasets as well as their respective content signatures or content hashes. Actual instances of crawl activities contain multiple registries (e.g., GBIF, iDigBio) and potentially thousands of datasets.
If you haven't yet tried Preston, please see the Installation section. Please also see a template repository and use cases for examples. If you are interested in learning how Preston works, please visit the architecture page.
- Usage - command available on the preston commandline tool
- Documentation
- Command Line Tool
update- update or track a biodiversity dataset graphls- list/print biodiversity dataset graphget- print biodiversity dataset graph node (e.g., dwca)history- show history of biodiversity dataset graph nodecopyTo- copies local versioned dataset graphs to another locationcheck/verify- verify/check the integrity of the locally versioned dataset graphs and their datasets.
- Use Cases
mining citationsweb access(nginx/caddy)archiving(rsync/archive.org/preston remote)data access monitorcompare versionsgenerating citationsfinding copies with hash-archive.orgtracking a GBIF IPTfinding text in tracked contentsgenerating publication using Jekyllparallel content tracking
- Prerequisites
- Install
- Building
- Contribute
- License
Preston was designed with the unix philosophy in mind: a simple tool with a specific focus that works well with others. For Preston, the focus is keeping track of biodiversity archives available through registries like GBIF, iDigBio and BioCASe. The functionality is currently available through a command line tool.
For documentation in man-pages style, see docs/preston.adoc.
The command line tool provides four commands: update, ls, get and history. In short, the commands are used to track and access DwC-A, EMLs and various registries. The output of the tools is nquads or tsv. Both output formats are structured in "columns" to form a three-term sentence per line. In a way, this output is telling you the story of your local biodiversity data graph in terms of simple sentences. This line-by-line format helps to re-use existing text processing tools like awk, sed, cut, etc. Also, tab-separated-values output plays well with spreadsheet applications and R.
The examples below assume that you've created a shortcut preston to java -jar preston.jar (see installation).
The update command updates your local biodiversity dataset graph using remote resources. By default, Preston uses GBIF, iDigBio and BioCASe to retrieve associated registries and data archives. The output is statements, expressed in nquads (or nquad-like tsv). An in-depth discussion of rdf, nquads and related topics are beyond the current scope. However, with a little patience, you can probably figure out what Preston is trying to communicate.
For instance:
$ preston update
<https://preston.guoda.bio> <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://www.w3.org/ns/prov#SoftwareAgent> .
<https://preston.guoda.bio> <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://www.w3.org/ns/prov#Agent> .
<https://preston.guoda.bio> <http://purl.org/dc/terms/description> "Preston is a software program that finds, archives and provides access to biodiversity datasets."@en .
<0b472626-1ef2-4c84-ab8f-9e455f7b6bb6> <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://www.w3.org/ns/prov#Activity> .
<0b472626-1ef2-4c84-ab8f-9e455f7b6bb6> <http://purl.org/dc/terms/description> "A crawl event that discovers biodiversity archives."@en .
<0b472626-1ef2-4c84-ab8f-9e455f7b6bb6> <http://www.w3.org/ns/prov#startedAtTime> "2018-09-05T04:42:40.108Z"^^<http://www.w3.org/2001/XMLSchema#dateTime> .
<0b472626-1ef2-4c84-ab8f-9e455f7b6bb6> <http://www.w3.org/ns/prov#wasStartedBy> <https://preston.guoda.bio> .
<0659a54f-b713-4f86-a917-5be166a14110> <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://www.w3.org/ns/prov#Entity> .
...tells us that there's a software program called "Preston" that started a crawl on 2018-09-05 . A little farther down, you'll see things like:
<https://gbif.org> <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://www.w3.org/ns/prov#Organization> .
<https://api.gbif.org/v1/dataset> <http://purl.org/dc/terms/description> "Provides a registry of Darwin Core archives, and EML descriptors."@en .
<https://api.gbif.org/v1/dataset> <http://purl.org/pav/createdBy> <https://gbif.org> .
<https://api.gbif.org/v1/dataset> <http://purl.org/dc/elements/1.1/format> "application/json" .
<https://api.gbif.org/v1/dataset> <http://purl.org/pav/hasVersion> <hash://sha256/5d1bb4f3a5a9da63fc76efc4d7b4a7debbec954bfd056544225c294fff679b4c> .which says that GBIF, an organization created a registry that has a version at hash://sha256/5d1bb4f3a5a9da63fc76efc4d7b4a7debbec954bfd056544225c294fff679b4c . This weird looking url is a content-addressed hash. Rather than describing where things are (e.g., https://eol.org), content-addressed hashes describe what they contain.
If you don't want to download the entire biodiversity dataset graph (~60GB) onto your computer, you can also use GBIF's dataset registry search api as a starting point. For instance, if you run preston update "http://api.gbif.org/v1/dataset/suggest?q=Amazon&type=OCCURRENCE", you only get occurence datasets that GBIF suggests are related to the Amazon. If you track these suggested datasets, you might see something like:
<http://plazi.cs.umb.edu/GgServer/dwca/FFBEFF81FE1A9007FFDFFC38FFDCFF90.zip> <http://purl.org/dc/elements/1.1/format> "application/dwca" .
<hash://sha256/5cba2f513fee9e1811fe023d54e074df2d562b4169b801f15abacd772e7528f8> <http://www.w3.org/ns/prov#generatedAtTime> "2018-09-05T05:11:33.592Z"^^<http://www.w3.org/2001/XMLSchema#dateTime> .
<hash://sha256/5cba2f513fee9e1811fe023d54e074df2d562b4169b801f15abacd772e7528f8> <http://www.w3.org/ns/prov#wasGeneratedBy> <21de25a8-927f-49a1-99be-725f1f506232> .
<http://plazi.cs.umb.edu/GgServer/dwca/FFBEFF81FE1A9007FFDFFC38FFDCFF90.zip> <http://purl.org/pav/hasVersion> <hash://sha256/5cba2f513fee9e1811fe023d54e074df2d562b4169b801f15abacd772e7528f8> .which tells us that a darwin core archive was found and a copy of it was made on 2018-09-05. The copy, or version, has a content hash of hash://sha256/5cba2f513fee9e1811fe023d54e074df2d562b4169b801f15abacd772e7528f8 . Incidentally, you can reach this same exact dataset at web-accessible preston archive. With this, we established that on 2018-09-05 a specific web addressed produced a specific content. On the next update run, Preston will download the content again. If the content is the same as before, nothing happens. If the content changed, a new version will be created associated with the same address, establishing a versioning of the content produced by the web address. This is addressed in a statement like <some hash> <.../previousVersion> <some previous hash>.
So, in a nutshell, the update process produces a detailed record of which resources are downloaded, what they look like and were they came from. You can retrieve the record of a successful run by using ls.
ls print the results of the previous updates. An update always refers to a previous update, so that a complete history can be printed / replayed of all past updates. So, the ls commands lists your (local) copy of the biodiversity dataset graph.
get retrieves a specific node in the biodiversity dataset graph. This can be a darwin core archive, EML file but also a copy of the iDigBio publisher registry. For instance, if you'd like to retrieve the node with DwC-A content, get the file and list the content using unzip and access the references in the taxa.txt file.
$ preston get hash://sha256/5cba2f513fee9e1811fe023d54e074df2d562b4169b801f15abacd772e7528f8 > dwca.zip
$ unzip -l dwca.zip
Archive: bla.zip
Length Date Time Name
--------- ---------- ----- ----
11694 2016-01-03 13:36 meta.xml
4664 2016-01-03 13:36 eml.xml
5533 2017-06-20 02:39 taxa.txt
284 2017-06-20 02:39 occurrences.txt
16978 2017-06-20 02:39 description.txt
54 2017-06-20 02:39 distribution.txt
48439 2017-06-20 02:39 media.txt
9280 2017-06-20 02:39 references.txt
33 2017-06-20 02:39 vernaculars.txt
--------- -------
96959 9 files
$ unzip -p dwca.zip taxa.txt | cut -f16
references
http://treatment.plazi.org/id/038787F9FE149009FED7FE39FEA9FCEE
http://treatment.plazi.org/id/038787F9FE1B9004FED7FA49FE99FA06
http://treatment.plazi.org/id/038787F9FE1E9002FED7FA5EFC1FFE3EThe implication of using content addressed storage is that if the hash is the same, you are guaranteed that the content is the same. So, you can reproduce the exact same results above if you have a file with the same content hash.
History helps to list your local content versions associated with a web address. Because the internet today might not be the internet of yesterday, and because publishers update their content for various reasons, Preston helps you keep track of the different versions retrieved from a particular location. Just like the Internet Archive's Way Back Machine keeps track of web page content, Preston helps you keep track of the datasets that you are interested in.
To inspect the history you can type:
$ preston history
<0659a54f-b713-4f86-a917-5be166a14110> <http://purl.org/pav/hasVersion> <hash://sha256/c253a5311a20c2fc082bf9bac87a1ec5eb6e4e51ff936e7be20c29c8e77dee55> .
<hash://sha256/b83cf099449dae3f633af618b19d05013953e7a1d7d97bc5ac01afd7bd9abe5d> <http://www.w3.org/ns/prov#generatedAtTime> "2018-09-04T20:48:35.096Z" .
<hash://sha256/b83cf099449dae3f633af618b19d05013953e7a1d7d97bc5ac01afd7bd9abe5d> <http://purl.org/pav/previousVersion> <hash://sha256/c253a5311a20c2fc082bf9bac87a1ec5eb6e4e51ff936e7be20c29c8e77dee55> .
<hash://sha256/7efdea9263e57605d2d2d8b79ccd26a55743123d0c974140c72c8c1cfc679b93> <http://www.w3.org/ns/prov#generatedAtTime> "2018-09-04T23:14:22.292Z" .
<hash://sha256/7efdea9263e57605d2d2d8b79ccd26a55743123d0c974140c72c8c1cfc679b93> <http://purl.org/pav/previousVersion> <hash://sha256/b83cf099449dae3f633af618b19d05013953e7a1d7d97bc5ac01afd7bd9abe5d> .
...By default, the history command shows the versions of your local biodiversity dataset graph as a whole. A list of versions associated with the sequence of updates. If you'd like to know what the UUID 0659a54f-b713-4f86-a917-5be166a14110 is described as, you can use ls and filter by the UUID:
$ preston ls | grep 0659a54f-b713-4f86-a917-5be166a14110
<0659a54f-b713-4f86-a917-5be166a14110> <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://www.w3.org/ns/prov#Entity> .
<0659a54f-b713-4f86-a917-5be166a14110> <http://purl.org/dc/terms/description> "A biodiversity dataset graph archive."@en .So, the UUID ending with 4110 is described as "A biodiversity dataset graph archive". This UUID is the same across all Preston updates, so in a way we are helping to create different versions of the same "a biodiversity dataset graph". Good to know right?
Preston stores versioned copies of biodiversity dataset graphs and their associated datasets in the data/ directory. The copyTo command moves the locally available biodiversity dataset graphs and their data to another location.
$ preston copyTo /home/someuser/target/data
indexing... done.
copying... [0.1]%
[...]
Copied [279636] datasets from [/home/someuser/source/data] to [/home/someuser/target/data] in [543] minutes.The check (aka verify) command takes the locally available versions of the dataset graph and verifies that the associated datasets are also available locally. In addition, the content hash (e.g., hash://sha256/...) for each local dataset graph and dataset is re-computed to verify that the content is still consistent with the content hash signatures recorded previously. The check command produces tab-separated values with five columns. The first column is the content hash of the file being checked, the second contains the location of the locally cached file, the third contains OK/FAIL to record the success of the check, the fourth gives a reason for check outcome and the fifth contains the total number of bytes of the local file associated with the hash.
$ preston check
hash://sha256/3eff98d4b66368fd8d1f8fa1af6a057774d8a407a4771490beeb9e7add76f362 file://some/path/3e/ff/3eff98d4b66368fd8d1f8fa1af6a057774d8a407a4771490beeb9e7add76f362 OK CONTENT_PRESENT_VALID_HASH 89931
hash://sha256/184886cc6ae4490a49a70b6fd9a3e1dfafce433fc8e3d022c89e0b75ea3cda0b file://some/path/18/48/184886cc6ae4490a49a70b6fd9a3e1dfafce433fc8e3d022c89e0b75ea3cda0b OK CONTENT_PRESENT_VALID_HASH 210344
...In the previous section the commands update, ls, get and history were introduced. Now, some use cases are covered to show how to combine these basic commands to make for useful operations. This is by no means an exhaustive list of all the potential uses, but instead is just to provide some inspiration on how to get the most out of preston.
The Ecological Metadata Language (EML) files contain citations, and your biodiversity dataset graph contains EML files. To extract all citations you can do:
# first make a list of all the emls
preston ls --log tsv | grep application/eml | cut -f1 > emls.txt
# then
preston ls -l tsv | grep -f emls.txt | grep "Version" | grep hash | cut -f3 | preston get | grep citation | sed 's/<[^>]*>//g' > citations.txt
head citations.txt
HW Jackson C, Ochieng J, Musila S, Navarro R, Kanga E (2018): A Checklist of the Mammals of Arabuko-Sokoke Forest, Kenya, 2018. v1.0. A Rocha Kenya. Dataset/Checklist. http://ipt.museums.or.ke/ipt/resource?r=asfmammals&v=1.0
Adda M., Sanou L., Bori H., 2018. Specimens d'herbier de la flore du Niger. Données d'occurrence publiées dans le cadre du Prjet BID Régional. CNSF-Niger
Michel.C., 2000. Arbres,arbustes et lianes des zones sèches d'Afrique de l'Ouest.3ème édition.Quae.MNHN.573p.
Hendrickson D A, Cohen A, Casarez M (2018): Ichthyology. v1.3. University of Texas at Austin, Biodiversity Collections. Dataset/Occurrence. http://ipt.tacc.utexas.edu/resource?r=tnhci&v=1.3
Urrutia N S (2014): Caracterización Florística de un Área Degradada por Actividad Minera en la Costa Caucana. v2.0. Instituto de Investigaciones Ambientales del Pacifico John Von Neumann (IIAP). Dataset/Occurrence. http://doi.org/10.15472/mkjqef
So, now we have a way to attribute each and every dataset individually.
Preston creates a "data" folder that stores the biodiversity datasets and associated information. For archiving, you can take this "data" folder, copy it and move it somewhere safe. You can also use tools like git-annex, rsync, or use distributed storage systems like the Interplanetary File System (ipfs) or Dat.
For instance, assuming that a preston data directory exists on a serverA which has ssh and rsync installed on it, you can keep a local copy in sync by running the following command on your local server:
$ rsync -Pavz preston@someserver:~/preston-archive/data /home/someuser/preston-archive/
where someserver is the remote server you'd like to sync with and /home/someuser/preston-archive is the place on your own server you'd like to store the rsync-ed Preston archive.
On a consumer internet connection with bandwidth < 10Mb/s, an initial sync with a remote trans-atlantic server with a 67GB preston archive took about 3 days. After the initial sync, only files that you don't have yet are included. For instance, if no new files are added to the remote preston archive, a sync take a few minutes instead of hours or days.
Note that ssh and rsync comes with frequently used linux distributions like Ubuntu v18.04 by default).
Alternatively, you can use an existing Preston remote (a publicly accessible Preston instance) to populate your local Preston installation using
$ preston cp --remote https://deeplinker.bio /some/local/path
indexing...
...Please note that depending on the size of the biodiversity datasets graph, this might take a while (hours, days, weeks depending on your network bandwidth). Unfortunately, at the time of writing, a progress monitor for the copy process is lacking.
The Internet Archive, a 501(c)(3) non-profit, is building a digital library of Internet sites and other cultural artifacts in digital form. Like a paper library, we provide free access to researchers, historians, scholars, the print disabled, and the general public. Our mission is to provide Universal Access to All Knowledge.
One of the services of the Internet Archive is the Wayback Machine . If your Preston archive is Web Accessible, you can use the Wayback Machine to make snapshots of your cached datasets. The bash scripts below can be used to do so.
#!/bin/bash
# Register all preston urls with internet-archive.org
#
set -x
domain="https:\/\/deeplinker\.bio"
function register_with_internet_archive {
zcat $1 | grep "hash:\/\/sha256" | sort | uniq | sed -e "s/hash:\/\/sha256/${domain}/g" | tee domain_urls.txt | sed -e 's/^/https:\/\/archive.org\/wayback\/available?url=/g' | xargs --no-run-if-empty -L1 curl -s | jq --raw-output ".archived_snapshots.closest | select(.available == true) | .url" | sort | uniq > domain_url_available_snapshots.txt
cat domain_url_available_snapshots.txt | sed -e "s/^.*${domain}//g" | sed -e "s/^/${domain}/g" > domain_urls_archived.txt
diff --changed-group-format='%>' --unchanged-group-format='' domain_urls_archived.txt domain_urls.txt > domain_urls_to_be_archived.txt
cat domain_urls_to_be_archived.txt | sed -e "s/^/https:\/\/web.archive.org\/save\//g" | tee domain_urls_save_request.txt | xargs --no-run-if-empty -L1 curl -s
}
/usr/local/bin/preston ls -l tsv | grep Version | head -n13 | cut -f1,3 | tr '\t' '\n' | grep -v "${domain}/\.well-known/genid" | sort | uniq | gzip > url_uniq.tsv.gz
register_with_internet_archive url_uniq.tsv.gzIn the script above, a list of urls is extracted and registered with archive.org if they haven't already.
Another way to submit content to the Internet Archive is using their s3-like interface via the Internet Archive Command-line Interface. Assuming that your preston archive is stored in /home/preston/preston-archive, your Internet Archive project id is preston-archive and that the commandline tool ia is configured properly, you can upload all the data using:
#!/bin/bash
find /home/preston/preston-archive | grep -v "tmp" | grep "data.*/data$" | sed 's/.*preston-archive\///g' | xargs -L1 bash -c 'echo upload preston-archive /home/preston/preston-archive/$0 --remote-name=$0' | tee uploaded_req.txt | xargs -L1 ia If you'd like to make your Preston archive accessible via http/https by using a nginx webserver, you can use a following address mapping to your nginx configuration:
location ~ "/\.well-known/genid/" {
return 302 https://www.w3.org/TR/rdf11-concepts/#section-skolemization;
}
location ~ "^/([0-9a-f]{2})([0-9a-f]{2})([0-9a-f]{2})([0-9a-f]{58})$" {
try_files /preston/$1/$2/$uri =404;
}The first location block redirects any URIs describing skolemized blank nodes to the appropriate w3c documentation on the topic. The second location block configures the server to attempt to retrieve a static file with a 64 hexadecimal sha256 hash from the appropriate data file in preston archive directory on the web server.
For more examples, please see the nginx configuration for the proxying the md5, sha1, and sha256 server deployments. At time of writing, these were the configurations used to power.
Similary, for Caddy, add the following to your Caddyfile:
redir 302 {
if {path} starts_with /.well-known/genid/
https://www.w3.org/TR/rdf11-concepts/#section-skolemization
}
rewrite {
r ^/([0-9a-f]{2})([0-9a-f]{2})([0-9a-f]{2})([0-9a-f]{58})$
to data/{1}/{2}/{path}
}Where you can replace data/ with the relative location of the local preston archive data directory.
With this, you can access your Preston archive remotely using the URLs like https://someserver/[sha256 content hash] . So, if you'd like to dereference (or download) hash://sha256/1846abf2b9623697cf9b2212e019bc1f6dc4a20da51b3b5629bfb964dc808c02 , you can now point your http client or browser at https://someserver/1846abf2b9623697cf9b2212e019bc1f6dc4a20da51b3b5629bfb964dc808c02 . Note that you do not need any other software than the (standard) nginx webserver, because you are serving the content as static files from the file system of your server.
By running update periodically and checking for "blank", or "missing" nodes (see blank skolemization), you can make a list of the dataset providers that went offline or are not responding.
The example below was created on 2018-09-05 using biodiversity dataset graph with hash hash://sha256/7efdea9263e57605d2d2d8b79ccd26a55743123d0c974140c72c8c1cfc679b93.
$ preston ls -l tsv | grep "/.well-known/genid/" | grep "Version" | cut -f1,3 | tr '\t' '\n' | grep -v "/.well-known/genid/" | grep -v "hash" | sort | uniq -c | sort -nr | head -n10
21 http://187.32.44.123/ipt/eml.do?r=camporupestre-15plot-survey-sampling-itacolomi-lagoa076-checklist
21 http://187.32.44.123/ipt/eml.do?r=camporupestre-15plot-survey-sampling-itacolomi-calais107-checklist
12 http://187.32.44.123/ipt/eml.do?r=2014-10-12-ufv-leep-buriti-543b4b1b47b42
6 http://xbiod.osu.edu/ipt/eml.do?r=rome
6 http://xbiod.osu.edu/ipt/eml.do?r=osuc
6 http://xbiod.osu.edu/ipt/archive.do?r=rome
6 http://91.151.189.38:8080/viript/eml.do?r=Avena_herbarium1
3 http://xbiod.osu.edu/ipt/eml.do?r=ucfc
3 http://xbiod.osu.edu/ipt/eml.do?r=proctos
3 http://xbiod.osu.edu/ipt/eml.do?r=platysKeeping track of changes across a diverse consortium of data publishers is necessary for reproducible workflows and reliable results. As datasets change, Preston can help you give insights into what changed exactly. For instance, the GBIF dataset registry changes as datasets are added, updated or deprecated. Below is an example of two version of the https://api.gbif.org/v1/dataset endpoint, one from 2018-09-03 and the other from 2018-09-04. Using jq and diff in combination with preston get and preston history gives us a way to check and see what changed.
$ preston ls | grep https://api.gbif.org/v1/dataset
<https://api.gbif.org/v1/dataset> <http://purl.org/pav/hasVersion> <hash://sha256/184886cc6ae4490a49a70b6fd9a3e1dfafce433fc8e3d022c89e0b75ea3cda0b> .
<https://api.gbif.org/v1/dataset> <http://purl.org/pav/hasVersion> <hash://sha256/1846abf2b9623697cf9b2212e019bc1f6dc4a20da51b3b5629bfb964dc808c02> .
$ preston get hash://sha256/184886cc6ae4490a49a70b6fd9a3e1dfafce433fc8e3d022c89e0b75ea3cda0b | jq . > one.json
$ preston get hash://sha256/1846abf2b9623697cf9b2212e019bc1f6dc4a20da51b3b5629bfb964dc808c02 | jq . > two.json
$ diff one.json two.json
20c20
< "text": "Ali P A, Maddison W P, Zahid M, Butt A (2017). New chrysilline and aelurilline jumping spiders from Pakistan (Araneae, Salticidae). Plazi.org taxonomic treatments database. Checklist dataset https://doi.org/10.3897/zookeys.783.21985 accessed via GBIF.org on 2018-08-31."
---
> "text": "Ali P A, Maddison W P, Zahid M, Butt A (2017). New chrysilline and aelurilline jumping spiders from Pakistan (Araneae, Salticidae). Plazi.org taxonomic treatments database. Checklist dataset https://doi.org/10.3897/zookeys.783.21985 accessed via GBIF.org on 2018-09-03."
248c248Preston provides both a date and a content-based identifier for the datasets that you are using and the biodiversity dataset graph as a whole. Also, it produces the information in a format that is machine readable. This enables the automated generation of citations, for human or machine consumption, as evidenced by the reference to a particular version of the biodiversity dataset graph in the previous section.
So now, instead of a citation to a dataset like:
Levatich T, Padilla F (2017). EOD - eBird Observation Dataset. Cornell Lab of Ornithology. Occurrence dataset https://doi.org/10.15468/aomfnb accessed via GBIF.org on 2018-09-11.
A citation might look something like:
Levatich T, Padilla F (2017). EOD - eBird Observation Dataset. Cornell Lab of Ornithology. Occurrence dataset hash://sha256/29d30b566f924355a383b13cd48c3aa239d42cba0a55f4ccfc2930289b88b43c accessed at http://ebirddata.ornith.cornell.edu/downloads/gbiff/dwca-1.0.zip at 2018-09-02 with provenance hash://sha256/b83cf099449dae3f633af618b19d05013953e7a1d7d97bc5ac01afd7bd9abe5d .
The latter citation tells you exactly what file was used and where it came from. The former tells you that some eBird dataset was accessed via GBIF on a specific date and leaves it up to the reader to figure out exactly which dataset was used.
hash-archive.org is a project by Ben Trask, the same person who suggested to use hash uris to represent content hashes (e.g., hash://sha256/...). The hash archive keeps track of what content specific urls created using content hashes. To make the hash archive update the hash associated with a url, you can send a http get request in the form of https://hash-archive.carlboettiger.info/api/enqueue/[some url] . For example, to register a url that is known to host an DwC-A at http://zoobank.org:8080/ipt/eml.do?r=zoobank, you can click on https://hash-archive.carlboettiger.info/api/enqueue/http://zoobank.org:8080/ipt/eml.do?r=zoobank , or using curl like
curl https://hash-archive.carlboettiger.info/api/enqueue/http://zoobank.org:8080/ipt/eml.do?r=zoobankOn successful completion of the request, hash-archive.org returns something like:
{
"url": "http://zoobank.org:8080/ipt/eml.do?r=zoobank",
"timestamp": 1537269631,
"status": 200,
"type": "text/xml;charset=utf-8",
"length": 3905,
"hashes": [
"md5-zyn7V5JlXkrqxJILT8ZfGw==",
"sha1-SALPtju8vNii2S/Rt3R946iKc0g=",
"sha256-yBrrBSjo86D8U4mniRsigr4ijoTAtXZ2aSJlhcTa1sQ=",
"sha384-IguP+tlrYZ8QVBC86YIPxf/7CWFhU2HTzxI2DYLq40mo1dwcS5yn6qJb0SatWaUH",
"sha512-sl3/Qm7Jd965F+QLkxbp/Xdsv7ZwWX6HKDgpwXk3OLyOGWpgym1HBSOEhRtMiH2g7MZzwKjyEyL4PajQAinj"
]
}This response indicates that the hash archive has independently downloaded the EML url and calculated various content hashes. Now, you should be able to do to https://hash-archive.org/history/http://zoobank.org:8080/ipt/eml.do?r=zoobank , and see the history of content that this particular url has produced.
In short, hash-archive provides a way to check whether content produced by a url has changed. Also, it provides a way to lookup which urls are associated with a unique content hash.
The example (also see related gist) below shows how Preston was used to register biodiversity source urls as well as Preston web-accessible urls via https://deeplinker.bio (see Web Access).
#!/bin/bash
# Register all preston urls with hash-archive.org
#
# Please replace "deeplinker\.bio" instances below with you own escaped hostname of your Preston instance.
# see https://preston.guoda.bio on how to install preston
#
preston ls -l tsv | grep Version | cut -f1,3 | tr '\t' '\n' | grep -v "deeplinker\.bio/\.well-known/genid" | sort | uniq | sed -e 's/hash:\/\/sha256/https:\/\/deeplinker.bio/g' | sed -e 's/^/https:\/\/hash-archive.carlboettiger.info\/api\/enqueue\//g' | xargs -L1 curl If all web-accessible Preston instances would periodically register their content like this, https://hash-archive.carlboettiger.info could serve as a way to lookup a backup for an archive that you got from an archive url that is no longer active.
GBIF's Integrated Publishing Toolkit (IPT) helps to publish and register biodiversity datasets with GBIF. IPT provide a RSS feeds that lists publicly available collections/datasets. Using this RSS feed, Preston can track datasets of individual IPTs, such as GBIF Norway's IPT at https://data.gbif.no/ipt . You can find the RSS link at the bottom of the home page of the ipt. GBIF Norway's RSS feed is https://data.gbif.no/ipt/rss.do . Now, you can update/track the IPT using Preston by running:
preston update https://data.gbif.no/ipt/rss.do
By running this periodically, you can keep track of dataset changes and retain historic datasets in your Preston archive.
The grep (or match) command searches nodes in the biodiversity dataset graph for text that matches a specified pattern. For each match it finds, it outputs the text that was matched and its location, including the node it was found in and where to find the text inside the node. If the match command encounters compressed files (e.g., .gz files), it will first decompress them. Files inside file archives (e.g., zip files) will also be searched. If no search pattern is specified, the grep command searches for strings that looks like a URL.
Here's the basic idea of matching/grepping text in a preston archive:
$ preston ls | preston grep "[some regex]"For a more complicated example, the entire Biodiversity Heritage Library is searched for occurrences "Aves" (birds) and characters preceding and following it. Notice the pattern preston ls | preston match [some regex] with a regex of [ A-Za-z]+Aves[ A-Za-z]+. Also, note the remotes that point to various remote locations of the Biodiversity Heritage Library using the --remote option.
For more information, see Poelen, Jorrit H. (2019). A biodiversity dataset graph: BHL (Version 0.0.2) [Data set]. Zenodo. http://doi.org/10.5281/zenodo.3484555 .
# find all mentions of text mentioning Aves (birds) in Biodiversity Heritage Library
$ preston ls --remote https://zenodo.org/record/3484555/files,https://deeplinker.bio\
| preston grep --no-cache --remote https://deeplinker.bio,https://zenodo.org/record/3484555/files "[ A-Za-z]+Aves[ A-Za-z]+"\
| head
<urn:uuid:17087386-391d-4192-b6fc-9a79daf846c6> <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://www.w3.org/ns/prov#Activity> <urn:uuid:17087386-391d-4192-b6fc-9a79daf846c6> .
<urn:uuid:17087386-391d-4192-b6fc-9a79daf846c6> <http://www.w3.org/ns/prov#used> <hash://sha256/e0c131ebf6ad2dce71ab9a10aa116dcedb219ae4539f9e5bf0e57b84f51f22ca> <urn:uuid:17087386-391d-4192-b6fc-9a79daf846c6> .
<urn:uuid:17087386-391d-4192-b6fc-9a79daf846c6> <http://purl.org/dc/terms/description> "An activity that finds the locations of text matching the regular expression '[ A-Za-z]+Aves[ A-Za-z]+' inside any encountered content (e.g., hash://sha256/... identifiers)."@en <urn:uuid:17087386-391d-4192-b6fc-9a79daf846c6> .
<cut:hash://sha256/e0c131ebf6ad2dce71ab9a10aa116dcedb219ae4539f9e5bf0e57b84f51f22ca!/b217065-217087> <http://www.w3.org/ns/prov#value> " Subclass Aves Carinate" <urn:uuid:17087386-391d-4192-b6fc-9a79daf846c6> .
<cut:hash://sha256/e0c131ebf6ad2dce71ab9a10aa116dcedb219ae4539f9e5bf0e57b84f51f22ca!/b217166-217188> <http://www.w3.org/ns/prov#value> " Subclass Aves Carinate" <urn:uuid:17087386-391d-4192-b6fc-9a79daf846c6> .
...In the example above, two matches are Subclass Aves Carinate in content hash://sha256/e0c131ebf6ad2dce71ab9a10aa116dcedb219ae4539f9e5bf0e57b84f51f22ca, content retrieved from <https://www.biodiversitylibrary.org/data/item.txt> at byte ranges b217065-217087 and b217166-217188 .
Selecting the byte ranges using the unix tool cut can be done with:
$ preston cat --remote https://deeplinker.bio hash://sha256/e0c131ebf6ad2dce71ab9a10aa116dcedb219ae4539f9e5bf0e57b84f51f22ca\
| cut -z -b217065-217087
Subclass Aves Carinate
Alternative, you can use Preston's built in cut notation:
$ preston cat --no-progress --no-cache --remote https://deeplinker.bio 'cut:hash://sha256/e0c131ebf6ad2dce71ab9a10aa116dcedb219ae4539f9e5bf0e57b84f51f22ca!/b217065-217087'
Subclass Aves Carinate
See #75 and https://jhpoelen.nl/bees for examples on how to generate a static website from a Preston biodiversity dataset graph.
A single biodiversity data archive/graph can be constructed, or updated, using parallel processes.
For instance, if you'd like to track two separate web locations in parallel, you can using GNU's parallel to do:
$ echo -e "https://example.org/bigdata1\nhttps://example.org/bigdata2" | parallel -j2 --line-buffer preston track
<https://preston.guoda.bio> <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://www.w3.org/ns/prov#SoftwareAgent> <19974b7b-d88b-4ffb-aa17-e12153956b86> .
<https://preston.guoda.bio> <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://www.w3.org/ns/prov#Agent> <19974b7b-d88b-4ffb-aa17-e12153956b86> .
<https://preston.guoda.bio> <http://purl.org/dc/terms/description> "Preston is a software program that finds, archives and provides access to biodiversity datasets."@en <19974b7b-d88b-4ffb-aa17-e12153956b86> .
...where echo -e "https://example.org/bigdata1\nhttps://example.org/bigdata2" contains two lines with each one url, and parallel -j2 --line-buffer preston track launches two preston processes, one for each url.
On completion, each Preston process adds it's provenance log to the end of the preston archive version history. So, after each track process has completed, you'll find two extra versions added to the biodiversity graph using preston history.
Preston needs Java 8+.
Preston is a stand-alone java application, packaged in a jarfile. You can build you own (see building) or download a prebuilt jar at releases.
See Quick Start for installing standalone Preston.
If you'd like to run Preston inside a docker container so that you don't have to worry about installing/conflicting java dependencies use:
- download the image
wget https://github.com/bio-guoda/preston/releases/download/0.3.1/preston.image.tar - load the image
sudo docker load --input preston.image.tar - run a container, mapping a host volume onto the containers /data folder
sudo docker run -v [some absolute host dir]/data:/data bio.guoda/preston:0.3.1 - generation of preston updating / crawl messages like:
<https://preston.guoda.bio> <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://www.w3.org/ns/prov#SoftwareAgent> .
<https://preston.guoda.bio> <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://www.w3.org/ns/prov#Agent> .
<https://preston.guoda.bio> <http://purl.org/dc/terms/description> "Preston is a software program that finds, archives and provides access to biodiversity datasets."@en .
<a4accddb-bf8a-477f-aa6f-413281c8d650> <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://www.w3.org/ns/prov#Activity> .
...
If you'd like to run Preston as a service to periodically update, you can use a systemd service combined with a systemd timer, or perhaps using a Jenkins job. Both have advantages. The following example focuses on systemd.
Example of preston.service :
[Unit]
Description=Preston tracks biodiversity datasets.
[Service]
Type=oneshot
User=preston
WorkingDirectory=/var/lib/preston
ExecStart=/usr/local/bin/preston update
Example of preston.timer :
[Unit]
Description=Run Preston
[Timer]
OnCalendar=weekly
RandomizedDelaySec=86400
[Install]
WantedBy= multi-user.target
After installing the systemd service and timer, apply changes by sudo systemctl daemon-reload and enable timer using sudo systemctl enable preston.timer and sudo systemctl start preston.timer to start the timer.
See systemd for more information.
Please use maven version 3.3+.
- Clone this repository
- Run tests using
mvn test(optional). - Run
mvn package -DskipTeststo build (standalone) jar - Copy
preston/target/preston-[version]-jar-with-dependencies.jarto[some dir]/preston.jar - Generate Asciidoc documentation using
preston gen-manpage --outdir docs/or similar.
Preston is made available through a maven repository.
To include preston in your project, add the following sections to your pom.xml (or equivalent for sbt, gradle etc):
<repositories>
<repository>
<id>depot.globalbioticinteractions.org</id>
<url>https://depot.globalbioticinteractions.org/release</url>
</repository>
</repositories>
<dependencies>
<dependency>
<groupId>bio.guoda</groupId>
<artifactId>preston</artifactId>
<version>0.0.1</version>
</dependency>
</dependencies>
Usage: <main class> [command] [command options]
Commands:
ls list biodiversity dataset graph
Usage: ls [options]
Options:
-l, --log
log format
Default: nquads
Possible Values: [tsv, nquads, dots]
get get biodiversity node(s)
Usage: get node id (e.g., [hash://sha256/8ed311...])
update update biodiversity dataset graph
Usage: update [options] content URLs to update. If specified, the seeds
will not be used.
Options:
-i, --incremental
resume unfinished update
Default: false
-l, --log
log format
Default: nquads
Possible Values: [tsv, nquads, dots]
-u, --seed
starting points for graph discovery. Only active when no content
urls are provided.
Default: [https://idigbio.org, https://gbif.org, http://biocase.org]
history show history of biodiversity dataset graph
Usage: history [options] biodiversity resource locator
Options:
-l, --log
log format
Default: nquads
Possible Values: [tsv, nquads, dots]
version show version
Usage: version
Feel free to join in. All welcome. Open an issue!
This work is funded in part by grant NSF OAC 1839201 from the National Science Foundation.
A list of publications using Preston or exploring the idea of using content-based identifiers as a building block for reliably reference data and their provenance.
Elliott M.J., Poelen J.H., Fortes J.A.B. (2020). Toward Reliable Biodiversity Dataset References. Ecological Informatics. https://doi.org/10.1016/j.ecoinf.2020.101132 hash://sha256/136c3c1808bcf463bb04b11622bb2e7b5fba28f5be1fc258c5ea55b3b84f482c
Elliott M.J., Poelen, J.H. & Fortes, J.A.B. (2023) Signing data citations enables data verification and citation persistence. Sci Data. https://doi.org/10.1038/s41597-023-02230-y hash://sha256/f849c870565f608899f183ca261365dce9c9f1c5441b1c779e0db49df9c2a19d
Poelen, Jorrit H.; Schulz, Kayja; Trei, Kelli J.; Rees, Jonathan A. (2019). Finding Identification of Keys in the Biodiversity Heritage Library. Biodiversity Heritage Library (BHL) and Global Names (GN) Workshop. Champaign, Illinois. Zenodo. http://doi.org/10.5281/zenodo.3311815 . Status = PUBLISHED; Acknowledgement of Federal Support = Yes
Poelen, J. H. (2019) To connect is to preserve: on frugal data integration and preservation solutions. Society for Preservation of Natural History Collections (SPNHC) Annual Meeting. Chicago. https://doi.org/10.17605/OSF.IO/A2V8G. Status = PUBLISHED; Acknowledgement of Federal Support = Yes
Poelen, J. H. (2020). Global Biotic Interactions: Benefits of Pragmatic Reuse of Species Interaction Datasets. https://doi.org/10.17605/OSF.IO/9JT24
Elliott, M. J. (2020). Reliable Biodiversity Dataset References. iDigBio Communications Luncheon, 10 February 2020. https://doi.org/10.17605/OSF.IO/FTZ9B
Poelen, J. H., Boettiger, C. (2020). Reliable Data Use In R. 4th Annual Digital Data in Biodiversity Research, 1-3 June 2020. https://doi.org/10.17605/OSF.IO/VKJ9Q
Elliott, M. J., Poelen, J. H., Fortes, J. A. B. (2020). Reliable Dataset Identifiers Are Essential Building Blocks For Reproducible Research. 4th Annual Digital Data in Biodiversity Research, 1-3 June 2020. http://doi.org/10.17605/OSF.IO/AT4XE
Boettiger, C (2020). Content-based Identifiers for Iterative Forecasts: A Proposal. Data One Webinars. Accessible at https://www.dataone.org/webinars/iterative-forecasts/ .
Poelen, Jorrit H., & Best, Jason. (2023, May 31). Signed Biodiversity Data Packages: A Method to Cite, Verify, Mobilize, and Future Proof, Large Image Corpora. hash://sha256/eec2c116dd7434863fd3dfb30658beb49726560e55a40459c89020a7b570e14f hash://md5/a96f09d798262518014fefc77495c2d6. Zenodo. https://doi.org/10.5281/zenodo.7990927
Noyes, JS. (2019). Universal Chalcidoidea Database World Wide Web electronic publication. http://www.nhm.ac.uk/chalcidoids. hash://sha256/ec1760dc83dfb17df003ef5e626b965dd4403850bc58286ac59c7ef3a447e063 hash://md5/b89cbd6285133d5c25ad2aef819de388 . (0.1) [Data set]. Zenodo. https://doi.org/10.5281/zenodo.7844167
Poelen, Jorrit H. (2023). A biodiversity dataset graph: GBIF, iDigBio, BioCASe hash://sha256/450deb8ed9092ac9b2f0f31d3dcf4e2b9be003c460df63dd6463d252bff37b55 hash://md5/898a9c02bedccaea5434ee4c6d64b7a2 (0.0.4) [Data set]. Zenodo. https://doi.org/10.5281/zenodo.7651831
Big Bee Community, Poelen, Jorrit H., & Seltmann, Katja. (2022). Xylocopa sonorina - UCSB-IZC00012194 - Bee Library - 73e389aa-5886-4c48-8778-ba8932d1bd7e hash://sha256/96bfde1efa599e0e8e61de18b14d61dd308737f684950e4079c04e9bc0f33958 hash://md5/4940f68c84cffa4412f7ffb98bb255bd (0.0.3) [Data set]. Zenodo. https://doi.org/10.5281/zenodo.7114665
Poelen, Jorrit H., & Groom, Quentin. (2022). Preserved Specimen Records with Still Images Registered Across Biodiversity Data Networks in Period 2019-2022 hash://sha256/da7450941e7179c973a2fe1127718541bca6ccafe0e4e2bfb7f7ca9dbb7adb86 (0.0.1) [Data set]. Zenodo. https://doi.org/10.5281/zenodo.7032574
J.H. Poelen. A biodiversity dataset graph: Biodiversity Heritage Library (BHL) hash://sha256/34ccd7cf7f4a1ea35ac6ae26a458bb603b2f6ee8ad36e1a58aa0261105d630b1, https://archive.org/details/preston-bhl (2019)
J.H. Poelen Biodiversity Dataset Archive. hash://sha256/8aacce08462b87a345d271081783bdd999663ef90099212c8831db399fc0831b, https://archive.org/details/biodiversity-dataset-archives (2019)
J.H. Poelen. A biodiversity dataset graph: DataONE. hash://sha256/2b5c445f0b7b918c14a50de36e29a32854ed55f00d8639e09f58f049b85e50e3, https://archive.org/details/preston-dataone (2019)
J.H. Poelen. A biodiversity dataset graph: BHL. hash://sha256/34ccd7cf7f4a1ea35ac6ae26a458bb603b2f6ee8ad36e1a58aa0261105d630b1, https://doi.org/10.5281/zenodo.3849560 (2020)
J.H. Poelen. A biodiversity dataset graph: DataONE. hash://sha256/2b5c445f0b7b918c14a50de36e29a32854ed55f00d8639e09f58f049b85e50e3, https://doi.org/10.5281/zenodo.3849494 (2020)
J.H. Poelen. A biodiversity dataset graph: GBIF, iDigBio, BioCASe. hash://sha256/8aacce08462b87a345d271081783bdd999663ef90099212c8831db399fc0831b, https://doi.org/10.5281/zenodo.3852671 (2020)
A biodiversity dataset graph: https://jhpoelen.nl/bats. 2021. hash://sha256/5150f699411c4433b0a6c111f8e6ec7fbae2c336ab237f3638bbdc9d0b2dda0d
A biodiversity dataset graph: https://jhpoelen.nl/rats. 2021. hash://sha256/812da92d28f6abbd8b26be507168877ede7dfd78f7cc5b79b417316cf64ff78c
A biodiversity dataset graph: https://jhpoelen.nl/bees. 2020. hash://sha256/85138e506a29fb73099fb050372d8a379794ab57fe4bfdf141743db0de2b985c
All software and scripts written for the purposes of this project are publicly available and released under open source licenses on GitHub within the following online repositories:
https://github.com/bio-guoda/preston
https://github.com/bio-guoda/preston-scripts
https://github.com/cboettig/contentid
https://discourse.gbif.org/t/finding-a-gbif-occurrence-from-a-specimen-code/3852/6
https://discourse.gbif.org/t/darwin-core-half-million-update/3652/10
https://discourse.gbif.org/t/gbif-exports-as-public-datasets-in-cloud-environments/1835/5
https://discourse.gbif.org/t/version-control-of-a-dataset/2633/4
https://discourse.gbif.org/t/when-to-assign-a-new-doi-to-an-existing-dataset/2319/3
https://discourse.gbif.org/t/toward-reliable-biodiversity-dataset-references/1637
https://discourse.gbif.org/t/10-transactional-mechanisms-and-provenance/2667/13
https://discourse.gbif.org/t/10-transactional-mechanisms-and-provenance/2667/51
https://discourse.gbif.org/t/searching-on-catalogue-number/3202/4?u=jhpoelen
ArctosDB/arctos#3950 (re: internet no-fly list - internet addresses blacklisted from image retrieval)
Response to NSF 20-015, Dear Colleague Letter:Request for Information on Data-Focused Cyberinfrastructure Needed to Support Future Data-Intensive Science and Engineering Research - https://www.nsf.gov/cise/oac/datacirfi/pdf/11202805209_Poelen.pdf
Vines, T. H., Albert, A. Y. K., Andrew, R. L., Débarre, F., Bock, D. G., Franklin, M. T., Gilbert, K. J., Moore, J.-S., Renaut, S., & Rennison, D. J. (2014). The Availability of Research Data Declines Rapidly with Article Age. Current Biology, 24(1), 94–97. https://doi.org/10.1016/j.cub.2013.11.014