HealthChain is an open-source SDK for production-ready healthcare AI. Skip months of custom integration work with built-in FHIR support, real-time EHR connectivity, and deployment tooling for healthcare AI/ML systems — all in Python.
pip install healthchainHealthChain is the quickest way for AI/ML engineers to integrate their models with real healthcare systems.
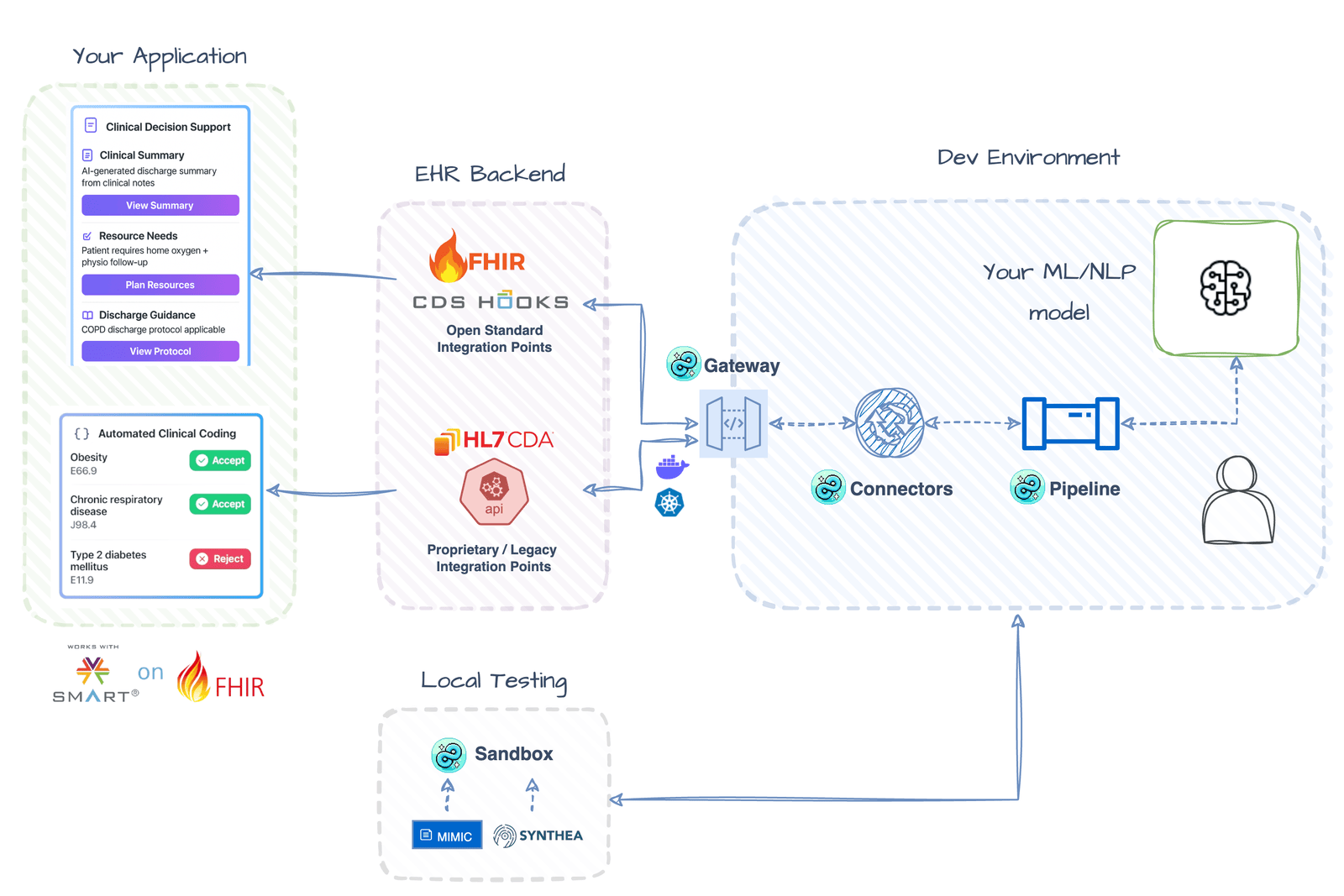
⚡️ Real-Time Clinical Workflow Integration Build CDS alerts and automated coding tools that integrate directly into Epic workflows Getting Started → |
🔥 FHIR Development Utilities Accelerate development with type-safe FHIR resource creation, validation helpers, and sandbox environments for testing clinical workflows Getting Started → |
🔌 Multi-EHR Data Aggregation Aggregate patient data from multiple FHIR sources into unified records with built-in NLP processing and automatic deduplication Getting Started → |
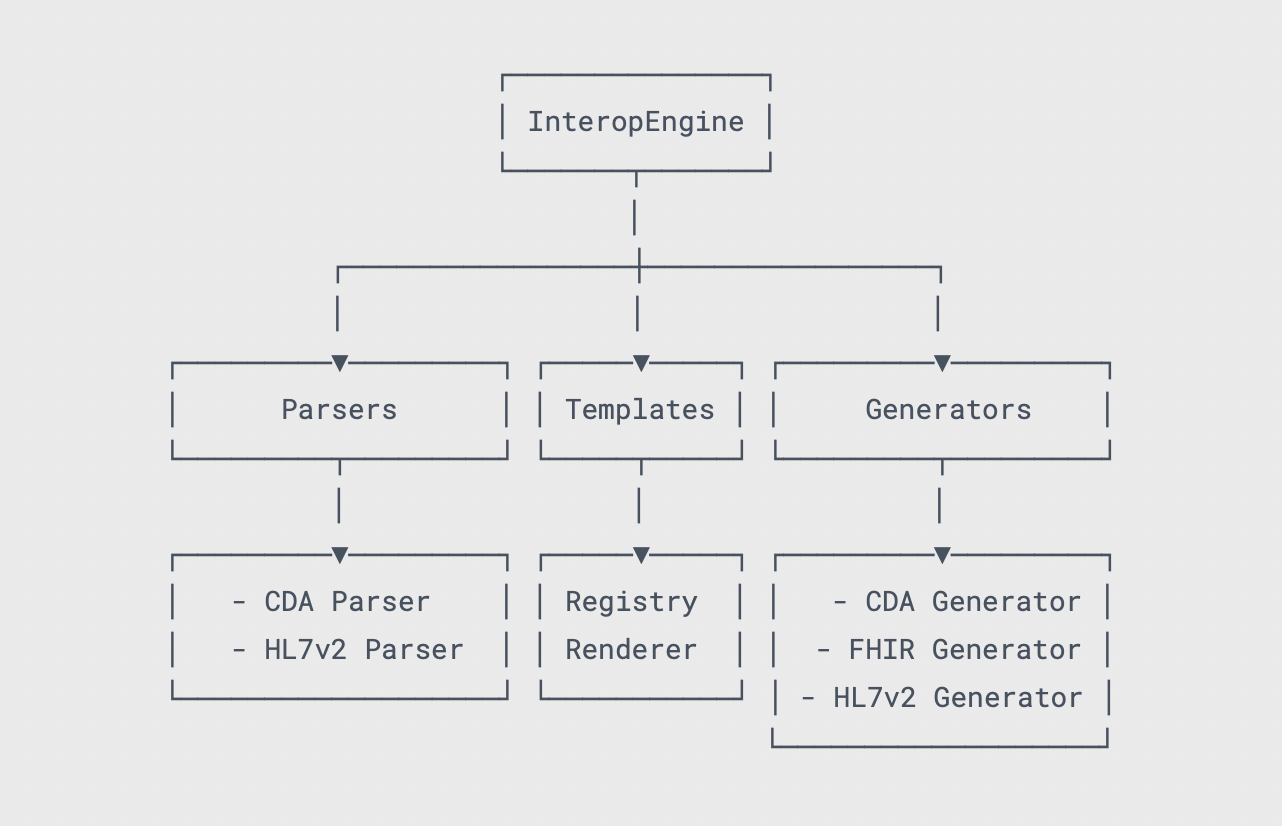
🔄 Healthcare Data Format Conversion Convert between FHIR and CDA formats with configuration-driven templates for unified data processing workflows Getting Started → |
|
🚀 Deploy ML Models as Healthcare APIs Turn any trained model into a production-ready FHIR endpoint with OAuth2 authentication and type-safe healthcare data handling Getting Started → |
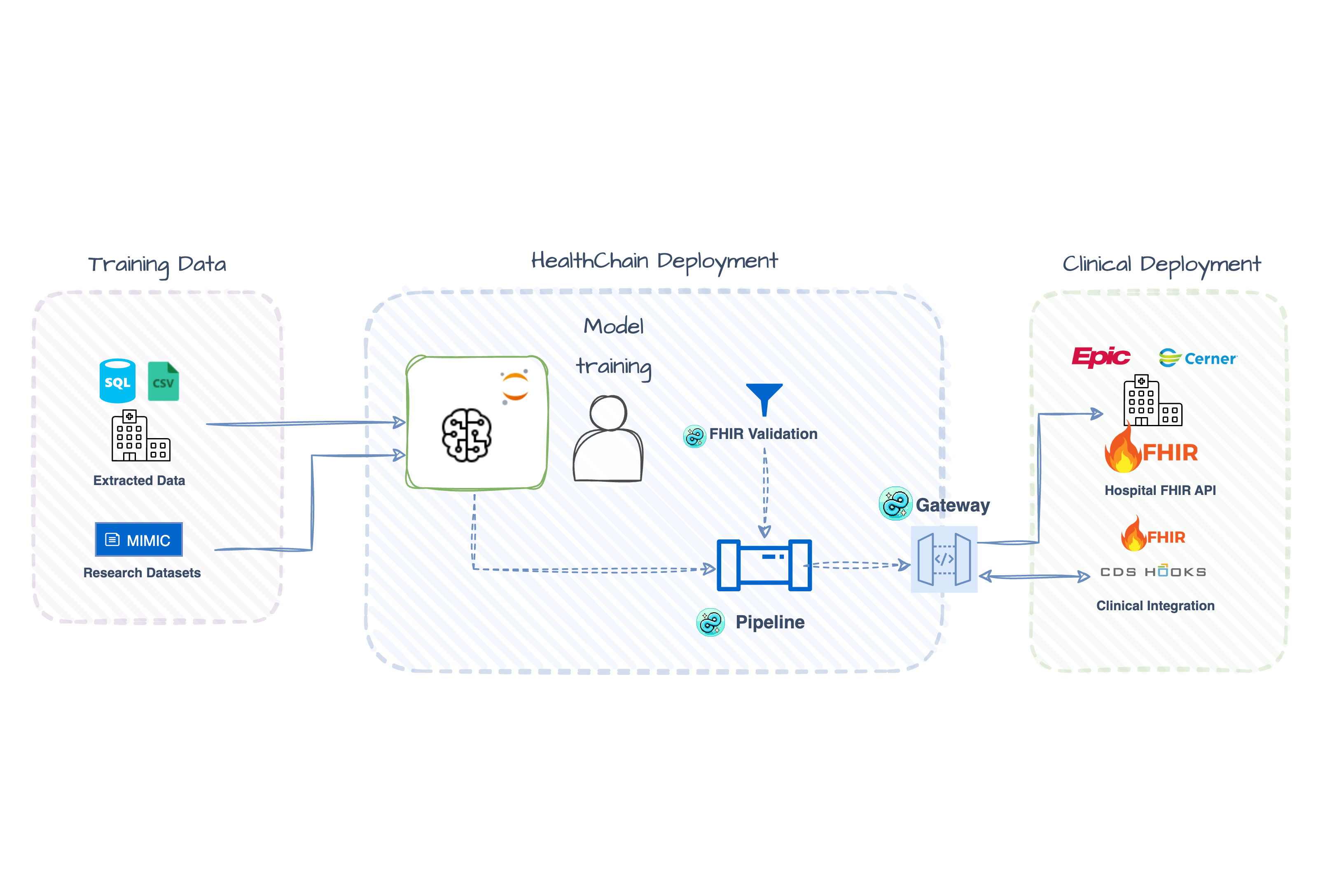
Electronic health record (EHR) data is specific, complex, and fragmented. Most healthcare AI projects require months of manual integration and custom validation on top of model development. This leads to fragile pipelines that break easily and consume valuable developer time.
HealthChain understands healthcare protocols and data formats natively, so you don't have to build that knowledge from scratch. Skip months of custom integration work and productionize your healthcare AI faster.
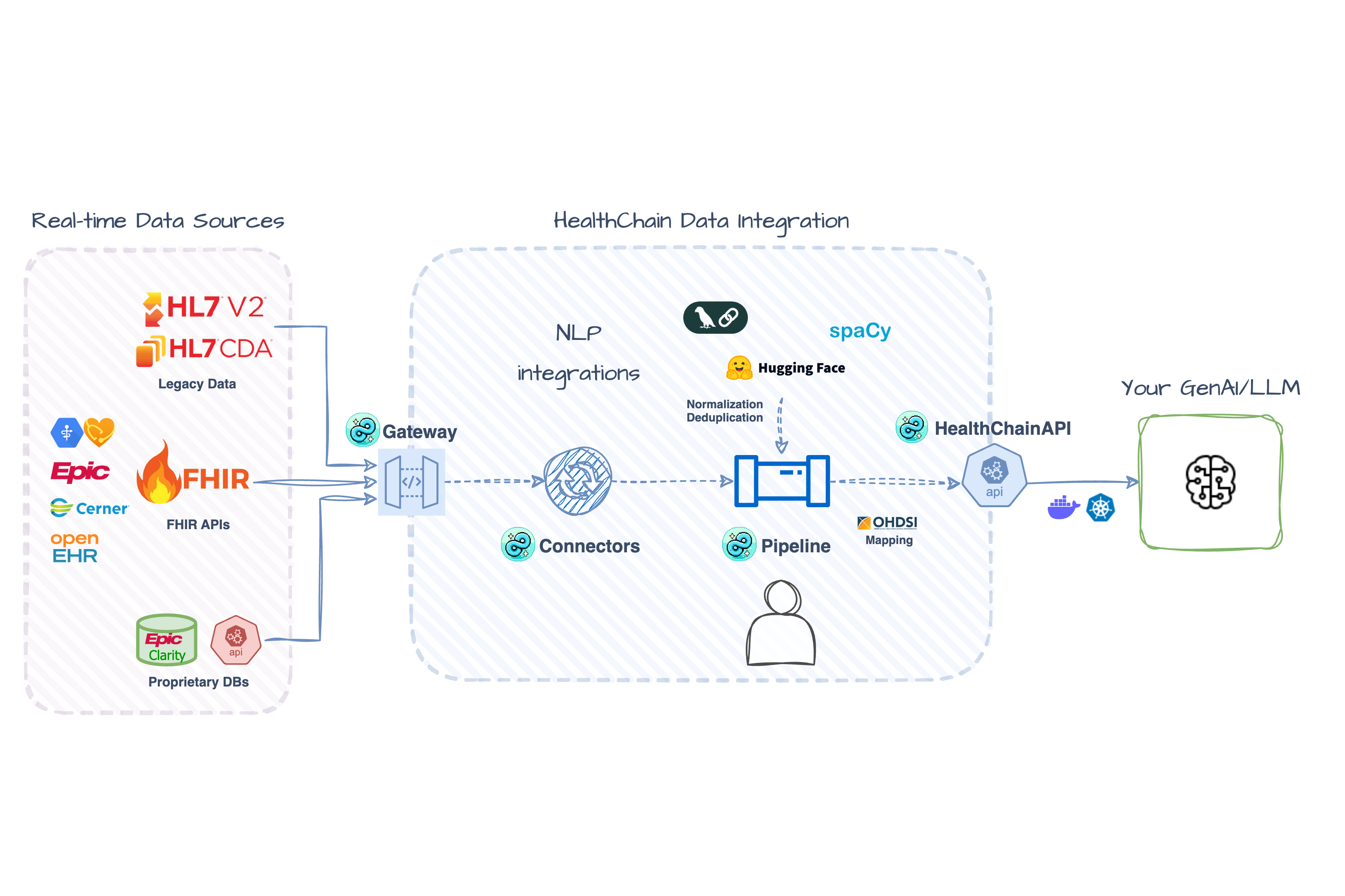
- Optimized for real-time - Connect to live FHIR APIs and integration points instead of stale data exports
- Automatic validation - Type-safe FHIR models prevent broken healthcare data
- Built-in NLP support - Extract structured data from clinical notes, output as FHIR
- Developer experience - Modular and extensible architecture works across any EHR system
- Production-ready - OAuth2-based authentication, Dockerized deployment, and structured audit hooks for real-world healthcare environments
Featured & Presented:
- Featured by Medplum for open source integration with Epic
- Featured in TLDR AI Newsletter (900K+ developers)
- Presented at NHS Python Open Source Conference (watch talk)
- Built from NHS AI deployment experience – read the origin story
Exploring HealthChain for your product or organization? Get in touch to discuss integrations, pilots, or collaborations, or join our Discord to connect with the community.
# Scaffold a new project
healthchain new my-app
cd my-app
# Run locally
healthchain serveSee the CLI reference for all commands.
Building a Pipeline [Docs]
from healthchain.pipeline import Pipeline
from healthchain.pipeline.components.integrations import SpacyNLP
from healthchain.io import Document
# Create medical NLP pipeline
nlp_pipeline = Pipeline[Document]()
nlp_pipeline.add_node(SpacyNLP.from_model_id("en_core_web_sm"))
nlp = nlp_pipeline.build()
doc = Document("Patient presents with hypertension and diabetes.")
result = nlp(doc)
spacy_doc = result.nlp.get_spacy_doc()
print(f"Entities: {[(ent.text, ent.label_) for ent in spacy_doc.ents]}")
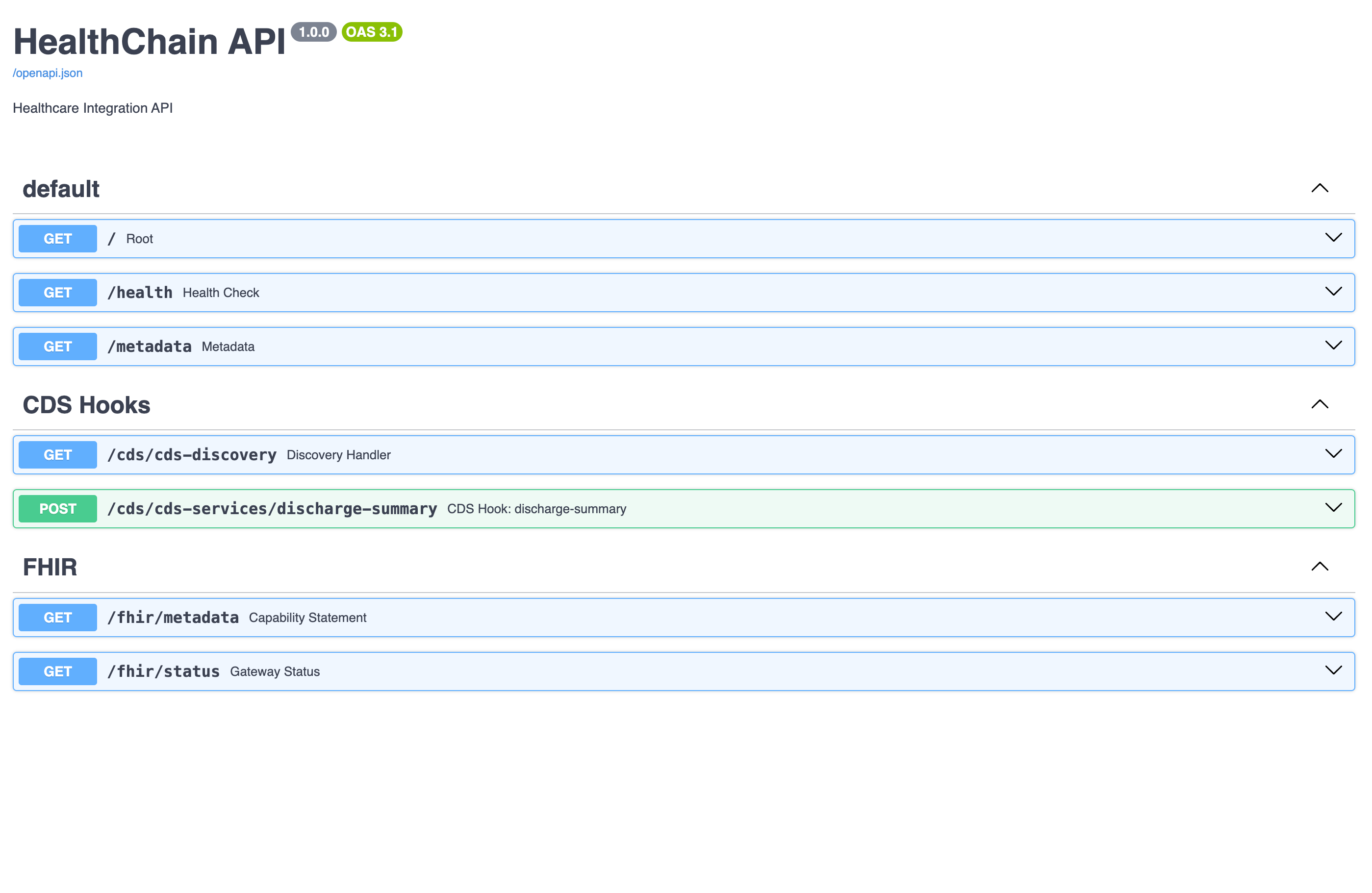
print(f"FHIR conditions: {result.fhir.problem_list}") # Auto-converted to FHIR BundleCreating a Gateway [Docs]
from healthchain.gateway import HealthChainAPI, FHIRGateway
from fhir.resources.patient import Patient
# Create healthcare application
app = HealthChainAPI(title="Multi-EHR Patient Data")
# Connect to multiple FHIR sources
fhir = FHIRGateway()
fhir.add_source("epic", "fhir://fhir.epic.com/r4?client_id=epic_client_id")
fhir.add_source("cerner", "fhir://fhir.cerner.com/r4?client_id=cerner_client_id")
@fhir.aggregate(Patient)
def enrich_patient_data(id: str, source: str) -> Patient:
"""Get patient data from any connected EHR and add AI enhancements"""
bundle = fhir.search(
Patient,
{"_id": id},
source,
add_provenance=True,
provenance_tag="ai-enhanced",
)
return bundle
app.register_gateway(fhir)
# Available at: GET /fhir/transform/Patient/123?source=epic
# Available at: GET /fhir/transform/Patient/123?source=cerner
if __name__ == "__main__":
import uvicorn
uvicorn.run(app, port=8000)Testing with Sandbox [Docs]
from healthchain.sandbox import SandboxClient
# Test CDS Hooks service with synthetic data
client = SandboxClient(
url="http://localhost:8000/cds/cds-services/discharge-summary",
workflow="encounter-discharge"
)
# Load from test datasets
client.load_from_registry(
"synthea-patient",
data_dir="./data/synthea",
resource_types=["Condition", "DocumentReference"],
sample_size=5
)
# Send requests and save results
responses = client.send_requests()
client.save_results("./output/")- 🔍 Data provenance and observability
- 🔒 Production security and compliance – enhanced authentication, audit logging, and HIPAA-aligned controls (encryption, access logging, configuration patterns)
- 🔄 HL7v2 parsing, FHIR profile conversion, and OMOP mapping
- 🧾 Da Vinci CRD/DTR/PAS prior authorization workflows
- 🚀 Kubernetes deployment and telemetry integration
- 📊 Model performance monitoring with MLFlow integration
- 🤖 MCP server integration
HealthChain is built for production healthcare systems. We prioritize contributors with:
- Healthcare product experience – shipped clinical systems, EHR integrations, or health data products
- FHIR expertise – designed or implemented FHIR APIs and interoperability solutions
- Healthcare security background – auth, privacy, compliance in regulated environments
If that's you, we'd love your input!
Get started:
- See the project board for current roadmap and active work
- Read CONTRIBUTING.md for guidelines and RFC process
- Discuss ideas in GitHub Discussions
- Join our Discord community
This project builds on fhir.resources and CDS Hooks standards developed by HL7 and Boston Children's Hospital.