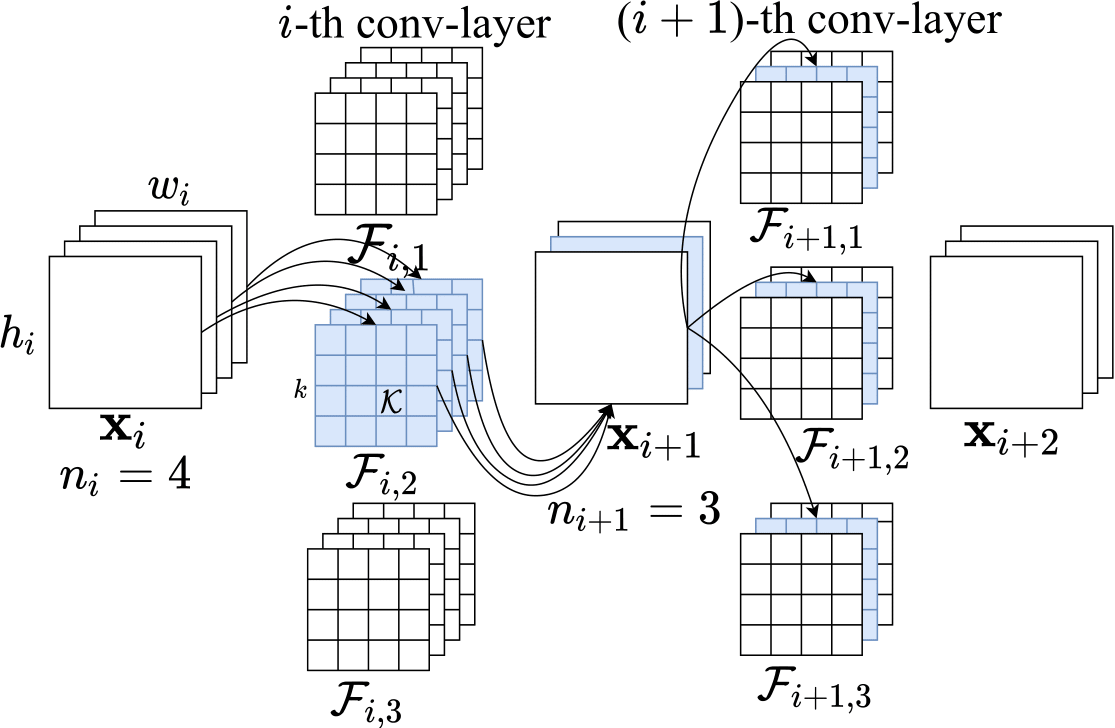
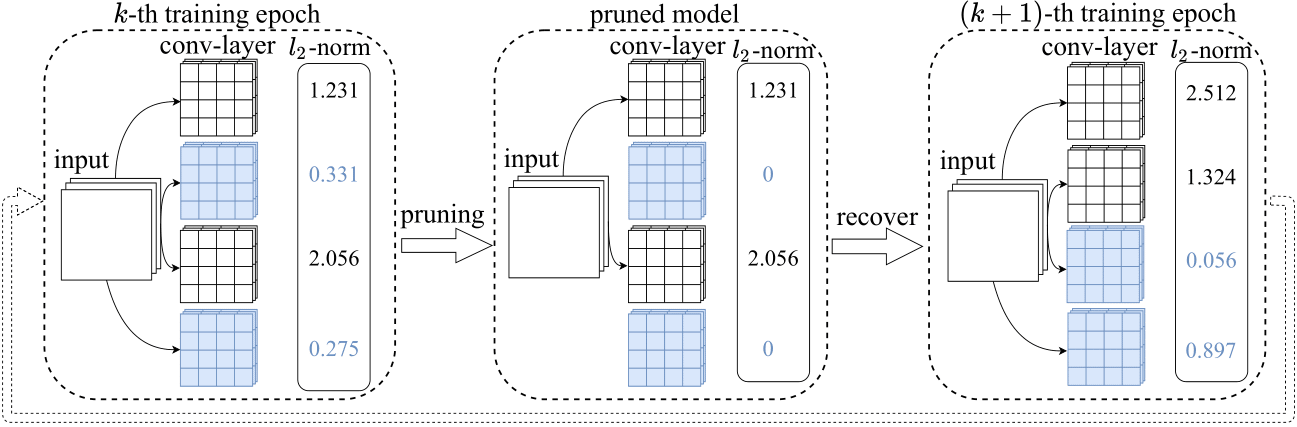
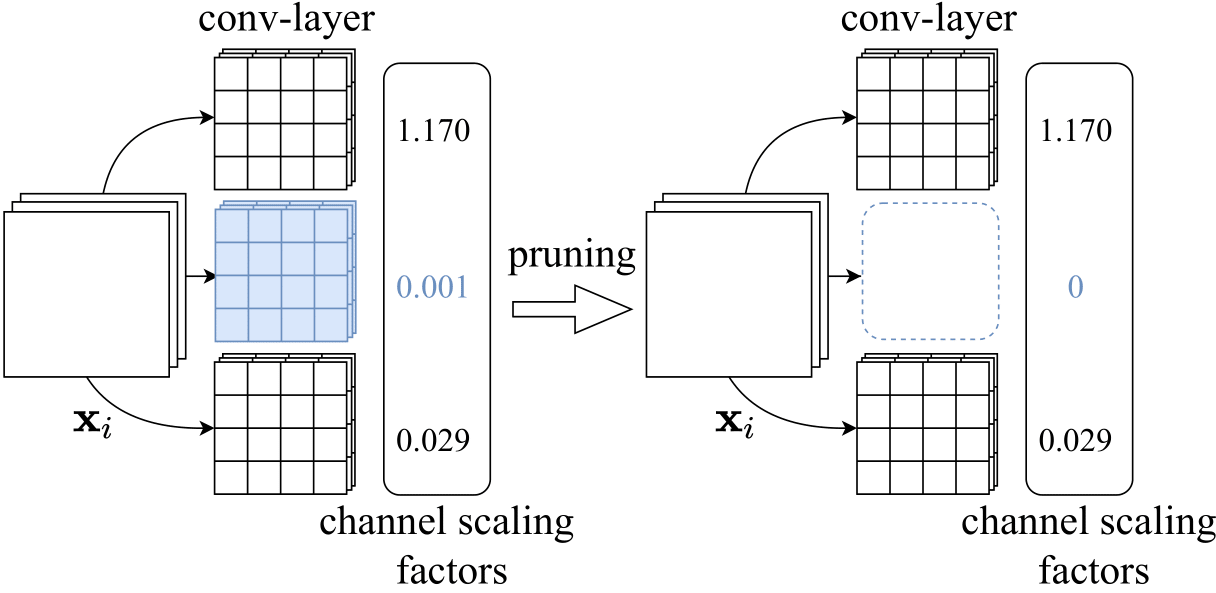
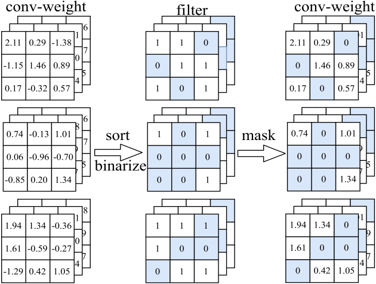
This is the implementation of our paper: Wenjie Rao, Sheng-hua Zhong "EEG Model Compression by Network Pruning for Emotion Recognition", accepted as a regular paper on International Joint Conference on Neural Networks (IJCNN)
With the development of deep learning on EEG-related tasks, the complexity of learning models has gradually increased. Unfortunately, the insufficient amount of EEG data limits the performance of complex models. Thus, model compression becomes an option to be seriously considered. So far, in EEG-related tasks, although some models used lightweight means such as separable convolution in their models, no existing work has directly attempted to compress the EEG model. In this paper, we try to investigate the state-of-the-art network pruning methods on commonly used EEG models for the emotion recognition task. In this work, we make several surprising observations that contradict common beliefs. Training a pruned model from scratch outperforms fine-tuning a pruned model with inherited weights, which means that the pruned structure itself is more important than the inherited weights. We can ignore the entire pruning pipeline and train the network from scratch using the predefined network architecture. We substantially reduce the computational resource overhead of the model while maintaining accuracy. In the best case, we achieve a 62.3% reduction in model size and a 64.3% reduction in computing operations without accuracy loss.
Please go to the working directory by:
$ cd ./code
Please create an anaconda virtual environment by:
$ conda create --name env1 python=3.8
Activate the virtual environment by:
$ conda activate env1
Install the requirements by:
$ pip3 install -r requirements.txt
To run the code for training the initial EEG model, please type the following command in the terminal:
$ python3 examples_fbccnn_unpruned.py where "fbccnn" can be replaced by other EEG models. The results will be saved into "result.txt" located in the same place as the script.
To model the EEG, please enter in the terminal
$ python3 fbccnn_LFP.pyLFP is one of the pruning methods and can be arbitrarily replaced with "NSFP/SFP/UWMP"
In the model recovery phase, enter
$ python3 examples_fbccnn_fine-tuned.pyWe provide two recovery methods: fine-tune and scratch.
LFP
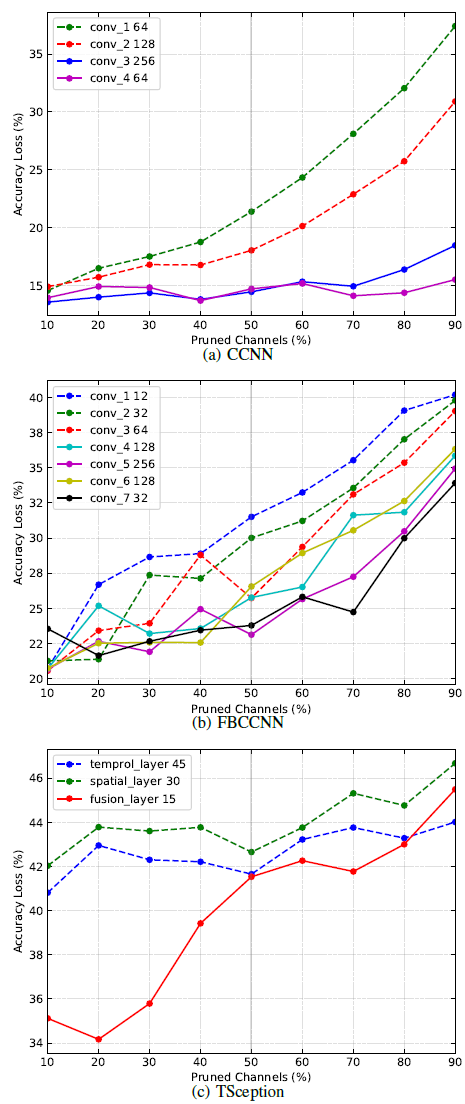
Different layers in the EEG model have different sensitivities to pruning. Pruning particular sensitive layers may result in a substantial loss of accuracy and are unrecoverable. Sensitivity analysis can be used to formulate pruning strategies to avoid excessive accuracy loss.
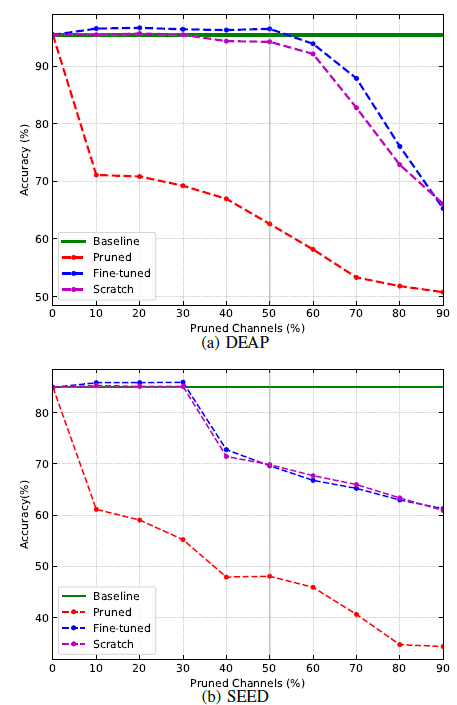
The pruning ratio is an essential parameter to determine the effectiveness of pruning. Too high a ratio will be destructive to the model, while too low a ratio can only save limited resources. We test the accuracy of TSception under various pruning ratios for two datasets.
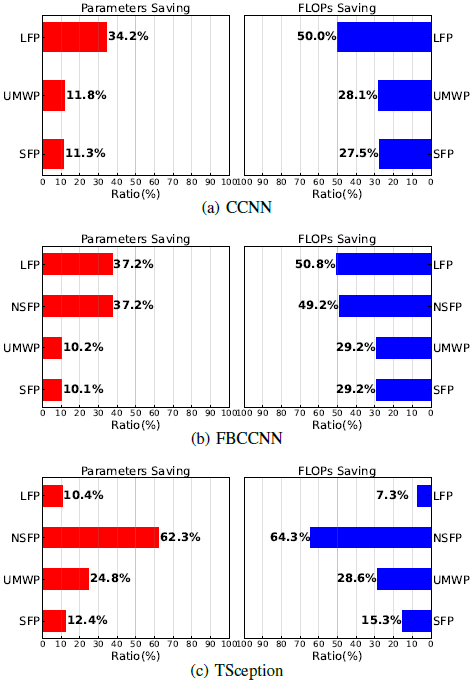
The efficiency of computational resource saving is an essential metric to evaluate a pruning algorithm. We compare the highest resource savings achieved by the four pruning methods without accuracy loss.
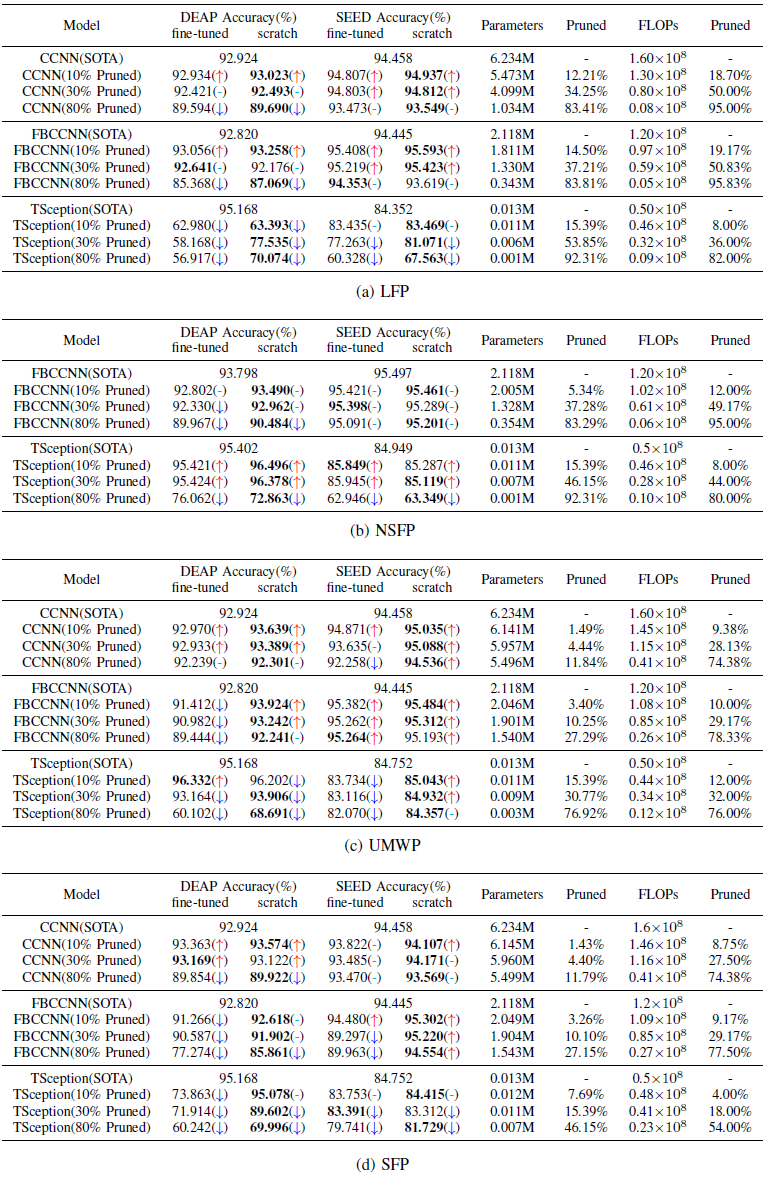
We compare the accuracy of network pruning and SOTA for three EEG models. As shown in Table, by comparing fine-tuned and scratched in columns 2-5.
We use both DEAP and SEED datasets in our experiments, please apply for download at the corresponding websites
- SEED dataset from Zheng et al.: Investigating critical frequency bands and channels for EEG-based emotion recognition with deep neural networks.
- DEAP dataset from Koelstra et al.: DEAP: A database for emotion analysis; using physiological signals.
Please place the "data_preprocessed_python" folder at the same location as the script
Import the relevant dataset (DEAP is used here) and pre-process the data:
from torcheeg.datasets import DEAPDataset
from torcheeg.datasets.constants.emotion_recognition.deap import DEAP_CHANNEL_LOCATION_DICT
dataset = DEAPDataset(io_path=f'./tmp_out/deap',
root_path='./tmp_in/data_preprocessed_python',
offline_transform=transforms.Compose(
[transforms.BandDifferentialEntropy(),
transforms.ToGrid(DEAP_CHANNEL_LOCATION_DICT)]),
online_transform=transforms.Compose([transforms.BaselineRemoval(),
transforms.ToTensor()]),
label_transform=transforms.Compose([
transforms.Select('valence'),
transforms.Binary(5.0),
]), num_worker=4)Defining the EEG model structure (CCNN here):
class CCNN(torch.nn.Module):
def __init__(self, in_channels=4, num_classes=3):
super().__init__()
self.conv1 = nn.Sequential(
nn.ZeroPad2d((1, 2, 1, 2)),
nn.Conv2d(in_channels, 64, kernel_size=4, stride=1),
nn.ReLU()
)
self.conv2 = nn.Sequential(
nn.ZeroPad2d((1, 2, 1, 2)),
nn.Conv2d(64, 128, kernel_size=4, stride=1),
nn.ReLU()
)
self.conv3 = nn.Sequential(
nn.ZeroPad2d((1, 2, 1, 2)),
nn.Conv2d(128, 256, kernel_size=4, stride=1),
nn.ReLU()
)
self.conv4 = nn.Sequential(
nn.ZeroPad2d((1, 2, 1, 2)),
nn.Conv2d(256, 64, kernel_size=4, stride=1),
nn.ReLU()
)
self.lin1 = nn.Linear(9 * 9 * 64, 1024)
self.lin2 = nn.Linear(1024, num_classes)
def forward(self, x):
x = self.conv1(x)
x = self.conv2(x)
x = self.conv3(x)
x = self.conv4(x)
x = x.flatten(start_dim=1)
x = self.lin1(x)
x = self.lin2(x)
return xSpecify the device and loss function used during training and test.
device = "cuda" if torch.cuda.is_available() else "cpu"
loss_fn = nn.CrossEntropyLoss()
batch_size = 64Use the training and validation scripts defined in the PyTorch tutorial:
def train(dataloader, model, loss_fn, optimizer):
size = len(dataloader.dataset)
model.train()
for batch_idx, batch in enumerate(dataloader):
X = batch[0].to(device)
y = batch[1].to(device)
# Compute prediction error
pred = model(X)
loss = loss_fn(pred, y)
# Backpropagation
optimizer.zero_grad()
loss.backward()
optimizer.step()
if batch_idx % 100 == 0:
loss, current = loss.item(), batch_idx * len(X)
print(f"loss: {loss:>7f} [{current:>5d}/{size:>5d}]")
def valid(dataloader, model, loss_fn):
size = len(dataloader.dataset)
num_batches = len(dataloader)
model.eval()
val_loss, correct = 0, 0
with torch.no_grad():
for batch in dataloader:
X = batch[0].to(device)
y = batch[1].to(device)
pred = model(X)
val_loss += loss_fn(pred, y).item()
correct += (pred.argmax(1) == y).type(torch.float).sum().item()
val_loss /= num_batches
correct /= size
print(f"Test Error: \n Accuracy: {(100*correct):>0.1f}%, Avg loss: {val_loss:>8f} \n")Traverse k folds and train the model separately for testing. We need to specify shuffle=True for the DataLoader of the training data set to avoid the deviation of the model training caused by consecutive labels of the same category.
for i, (train_dataset, val_dataset) in enumerate(k_fold.split(dataset)):
model = CNN().to(device)
optimizer = torch.optim.Adam(model.parameters(), lr=1e-4)
train_loader = DataLoader(train_dataset, batch_size=batch_size, shuffle=True)
val_loader = DataLoader(val_dataset, batch_size=batch_size, shuffle=False)
epochs = 50
for t in range(epochs):
print(f"Epoch {t+1}\n-------------------------------")
train(train_loader, model, loss_fn, optimizer)
valid(val_loader, model, loss_fn)
print("Done!")The author would like to thank ShenZhen University, National Natural Science Foundation of China
TorchEEG has a MIT license, as found in the LICENSE file.