| Health | Main Release (w/ Mantid Framework) | Dev Release (w/o Mantid Framework) |
|---|---|---|
 |
||
This project is trying to implement total scattering data reduction for neutron time-of-flight diffractometers using the algorithms currently available in the Mantid framework
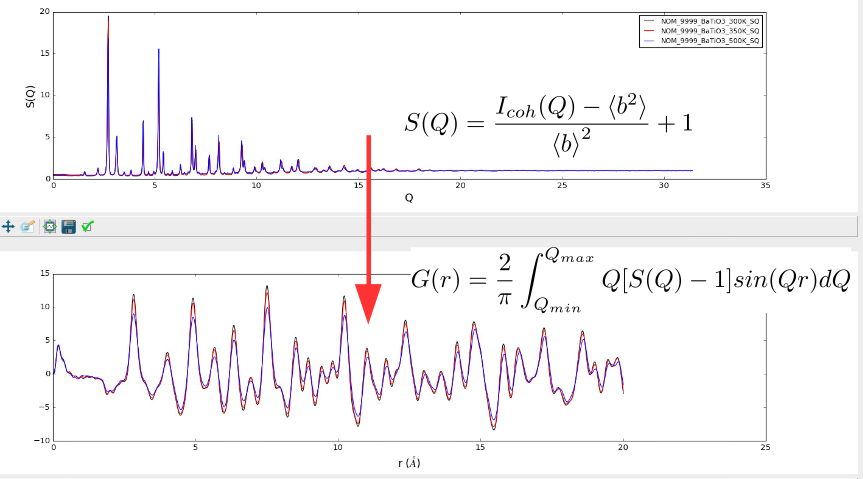
This entails taking raw neutron counts from detectors in the diffraction experiment and turning them into the reciprocal-space structure factor patterns, F(Q) or S(Q), and applying a Fourier Transform to real-space to give the pair distribution fuction, PDF.
This is the future backend for the ADDIE project and hopes to support multiple diffractometers performing total scattering measurements.
Running mantidtotalscattering will generate the total scattering data in
reciprocal space saved as NeXus file, if one is using the multiple bank mode.
Another alternative mode is the single bank mode which will merge spectra from
all detectors into a single pattern. The single bank mode will be taken in the
auto reduction implementation since otherwise manual efforts are needed to merge
the different banks data. When using the single bank mode, no Bragg data will be
saved. With the multiple bank mode, both the total scattering data and the Bragg
diffraction data will be saved. For the Bragg data output, there are two
available formats -- the GSS format and the XYE format. The GSS data file could
be loaded into the ADDIE interface (see the link above) for visualization. Here,
it is noteworthy that if one is loading in the GSS data in Mantid, one has to
rebin the loaded in workspace first (since the output GSS data is ragged, i.e.,
different banks are not sharing the common x-axis, for the purpose of removing
the non-sense data in each bank), followed by running the Mantid algorithm
ConvertToPointData.
Please follow the instructions provided at https://pixi.sh/latest/installation/
The following will clone the repo and instruct pixi to download and setup the dependencies in a sandboxed environment.
git clone https://github.com/neutrons/mantid_total_scattering.git
cd mantid_total_scattering
pixi installTo update
pixi updateCheat sheet for running local tasks with pixi,
pixi shell # activate the pixi shell, something analogous to `conda activate`
mantidtotalscattering input.json # the command is only available when pixi shell is active
pytest # run tests
flake8 total_scattering/ # format source codes with `flake8`
exit # exit the pixi shell
pixi run mantidtotalscattering input.json # run application task without activating pixi shell
pixi run pytest
pixi run flake8 total_scattering/
pixi run test # specifically defined test task
pixi run link # specifically defined link task
pixi shell -e local-mantid # activate the pixi shell with the `local-mantid` environmentconda install -c neutrons mantid-total-scatteringTo update
conda update -c neutrons mantid-total-scatteringIf you have an error (see below for example) related to the libGL library, you may not have it installed for the Mantid Framework to work. See instructions here for installing the necessary libraries for different OS
Example error:
ImportError: First import of "._api" failed with "libGL.so.1: cannot open shared object file...
To launch the total scattering script, complete the input JSON file (found in examples directory), and run:
pixi run mantidtotalscattering examples/sns/nomad_simple.jsonmantidtotalscattering examples/sns/nomad_simple.jsonPlease follow the directions provided [here[(https://developer.mantidproject.org/GettingStarted/GettingStartedCondaLinux.html#gettingstartedcondalinux)] to build a local version of mantid.
Once built, you must update some values in your pyproject.toml:
MANTID_BUILD_DIR: The path to your local mantid's build directory.MANTID_SRC_DIR: The path to your local mantid's repository root.
You may run mantidtotalscattering as normal via:
pixi run mantidtotalscattering-local examples/sns/nomad_simple.jsonOr you may launch mantidworkbench with the following command:
pixi run workbench-localThis will launch mantidworkbench in a seperate sandboxed pixi environement.
Then we can load the following script into mantidworkbench and execute it,
import json
from total_scattering.reduction import TotalScatteringReduction
from total_scattering.reduction import validateConfig
with open("FULL_PATH_TO_INPUT_JSON_FILE", "r") as handle:
config = json.load(handle)
# validate the config
validateConfig(config)
# Run total scattering reduction
TotalScatteringReduction(config)where we need to replace FULL_PATH_TO_INPUT_JSON_FILE with the full path to
our input json file.
Or, we can use the following script inside a general version of MantidWorkbench,
from mantid.simpleapi import *
import matplotlib.pyplot as plt
import numpy as np
mts_path = os.path.join(
"/gpfs/neutronsfs/instruments/NOM/shared/Dev/mantid_total_scattering"
)
sys.path.append(mts_path)
mts_path = os.path.join(
"/gpfs/neutronsfs/instruments/NOM/shared/Dev",
"mantid_total_scattering/.venv/lib/python3.10/site-packages"
)
sys.path.append(mts_path)
from total_scattering.reduction.total_scattering_reduction import \
TotalScatteringReduction # noqa: E407
from total_scattering.reduction.validator import validateConfig # noqa: E407
from total_scattering import __version__ as mts_version # noqa: E407
from total_scattering.reduction.normalizations import Material # noqa: E407
import json
with open("/SNS/PG3/IPTS-34378/shared/autoreduce/MTSRed/Input/PG3_59120.json", "r") as f:
config = json.load(f)
sofq = TotalScatteringReduction(config)To build and run the tests via pytest, use:
pixi run testN. B. This is assuming that the pixi environment mentioned above is installed.
git branch --track main origin/main # create a local main branch set to follow remote main
git checkout main
git fetch -p -t # fetch all changes from the remote repo
git rebase -v origin/main # sync with remote main branch
git merge --ff-only origin/qa # merge in all of the changes in branch next
git tag v1.0.7 # create the tag in the format that versioneer has been configured
git push origin v1.0.7 # push the tag to remote to kick off the deploy stepThe steps above will tag a new main release (the production release) and will
kick off the pipeline action to push the release to the neutrons Anaconda channel.
If we want to tag a release candidate, follow the steps below.
git branch --track qa origin/qa # create a local main branch set to follow remote main
git checkout qa
git fetch -p -t # fetch all changes from the remote repo
git rebase -v origin/qa # sync with remote main branch
git merge --ff-only origin/next # merge in all of the changes in branch next
git tag v1.0.8rc1 # create the tag in the format that versioneer has been configured
git push origin v1.0.8rc1 # push the tag to remote to kick off the deploy stepThis will kick off the pipeline action to push the release candidate to the neutrons Anaconda channel.
Releases will be deployed on the Analysis Cluster via a seperate deployment pipeline.