The most advanced and compact 3d engine ever implemented in DNA code.






- Synthesize the oligonucleotides from the cube3d.dna file.
- Arrange the test tubes as shown in the diagram below.
- Don't forget to provide the initial concentrations according to the table below.
- Use a pipette to encode the position (row and column) of each tube to start the computation.
Environment variables
q = 0.01
cxtm = 0.606
axp = 0.606
cytm = 0.898
ayp = 0.898
cztm = 1.243
azp = 1.243
mxyzm = 0.3
nx = 0.036 + 0.555 Col + 0.147 Row
ny = 0.853 + -0.517 Row
nz = 0.737 + -0.270 Col + 0.302 Row
- Pick the fluorophore of your favorite color and attach it to the
Strand R0, so that it activates when theRspecies are being produced. - Use a light source with a specific wavelength (depending on the fluorophore you've chosen) to render the result.
Disclaimer: perhaps it would not be a good idea to try this experiment in a real lab, because it will cost you a lot of money and most likely won’t work as intended the first time.
 Ray marching common implementation 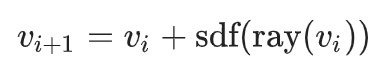
|
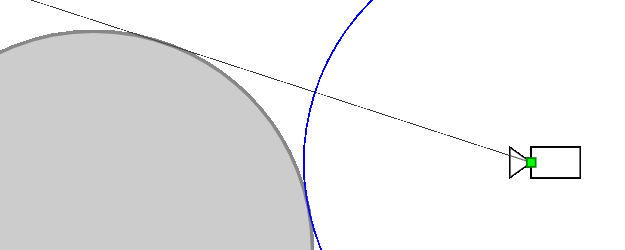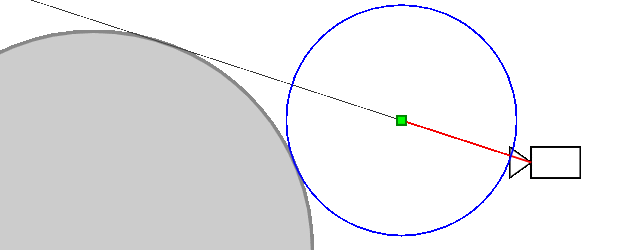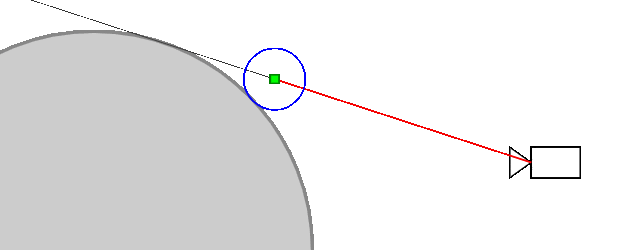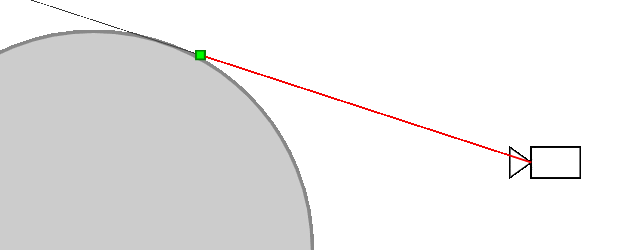 Ray marching differential form used here 
|
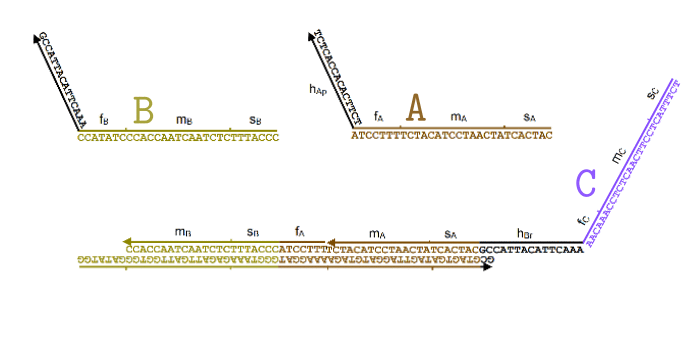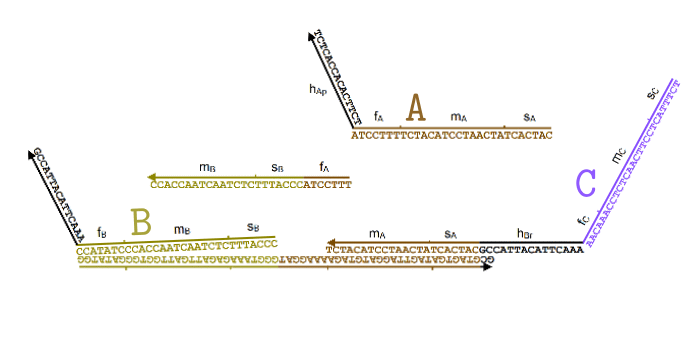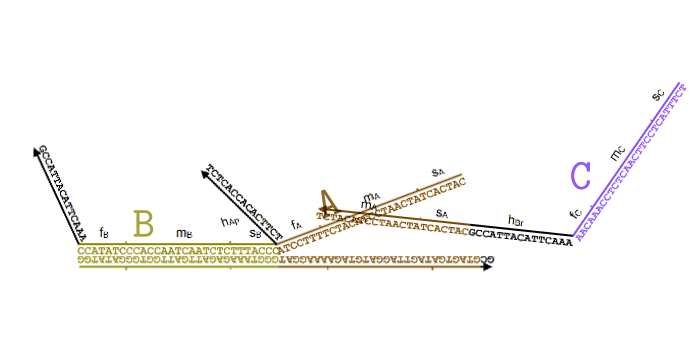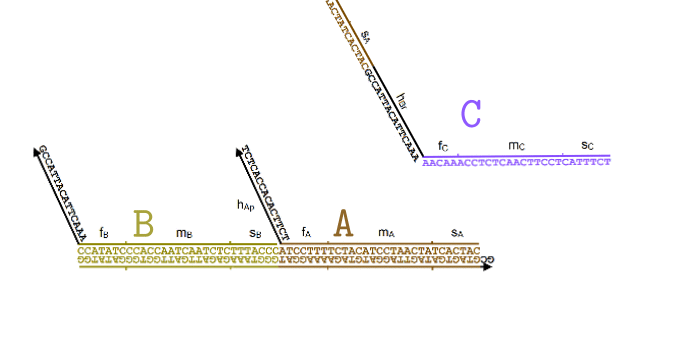 Simplified animation of the toehold mediated strand displacement technique (based on supplementary materials from [2]) |
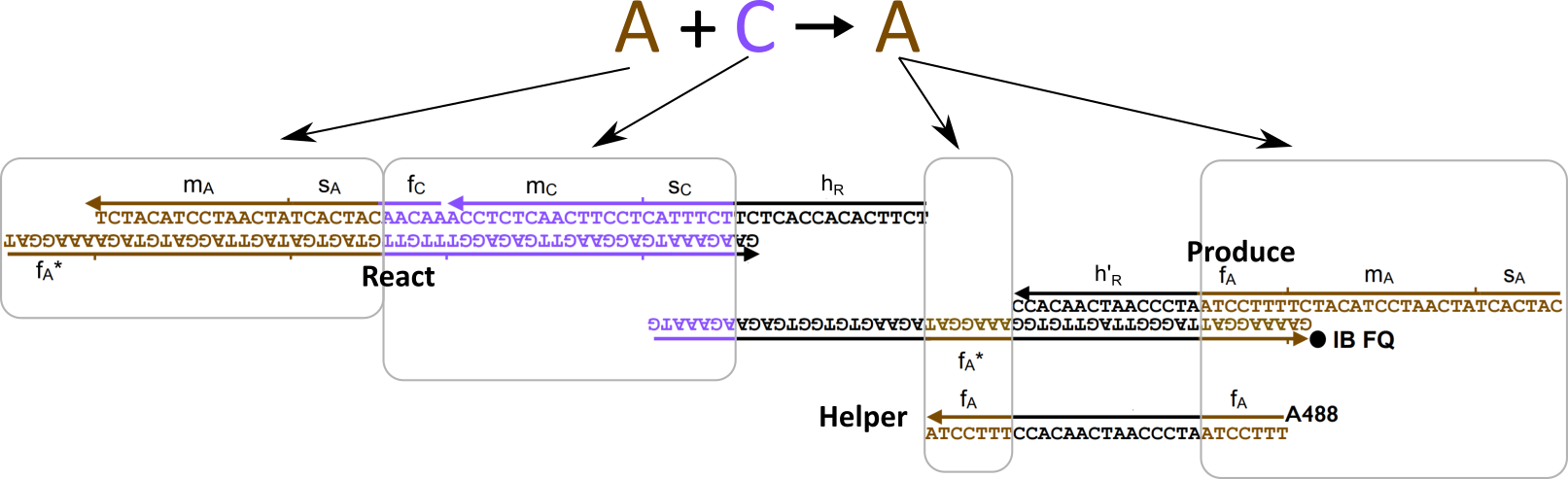 The types of oligonucleotides required for a single reaction (based on supplementary materials from [2]) |
 Data flow graph of the JS implementation. |
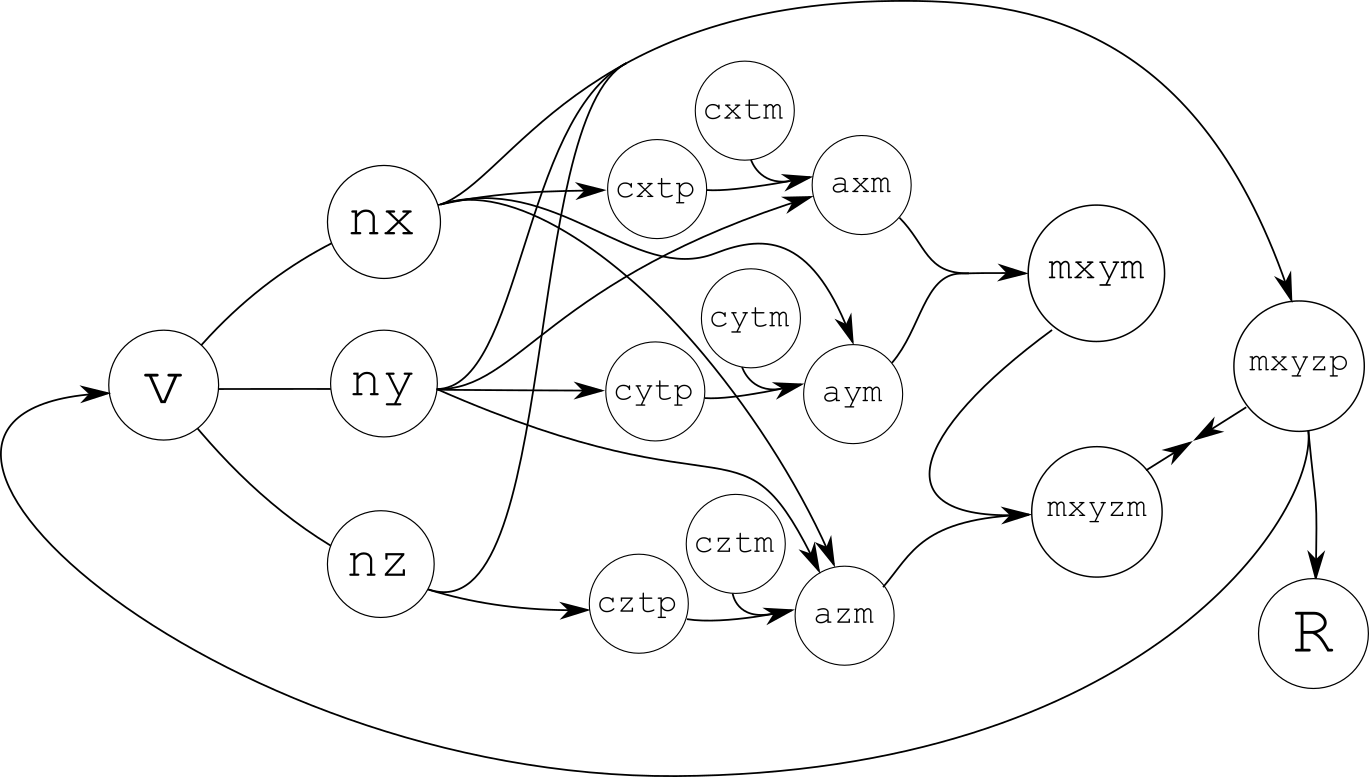 Species interaction graph of this implementation. |
 Unminified source code of this implementation which just comprises plain reactions with a few macros. The minifier was written in Wolfram Language specially for this project. |
 CRN++ source code for comparison (CRN++ is not used in this implementation). |
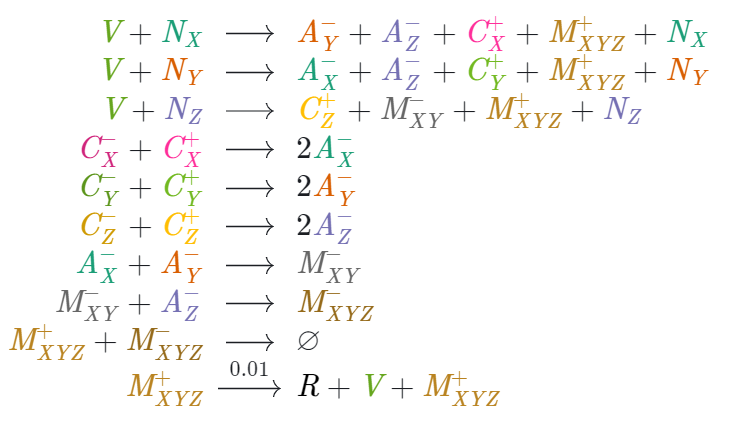 10 reactions after the minification used in this project. Since the piperine compiler doesn't support more than 2 products on the right-hand side, the reactions were split. |
 CRN++ 70 reactions output for comparison (CRN++ is not used in this implementation). |
- Install the piperine compiler by the DNA and Natural Algorithms Group.
- Run the following command
piperine-design cube3d.crn --maxspurious 0.765
- Wait 2-3 hours for the compilation results.
-
David Soloveichik, Georg Seelig and Erik Winfree
DNA as a universal substrate for chemical kinetics
Proceedings of the National Academy of Sciences Mar 2010, 107 (12) 5393-5398; DOI: 10.1073/pnas.0909380107 -
Niranjan Srinivas, James Parkin, Georg Seelig, Erik Winfree and David Soloveichik
Enzyme-free nucleic acid dynamical systems
Science 358, eaal2052 (2017).
Some images were taken from supplementary materials -
Chalk, Cameron, Niels Kornerup, Wyatt Reeves, and David Soloveichik.
Composable rate-independent computation in continuous chemical reaction networks.
International Conference on Computational Methods in Systems Biology, pp. 256-273. Springer, Cham, 2018. -
Chen, Ho-Lin, David Doty, and David Soloveichik.
Rate-independent computation in continuous chemical reaction networks.
Proceedings of the 5th conference on Innovations in theoretical computer science. 2014. -
Marko Vasic, David Soloveichik and Sarfraz Khurshid
CRN++: Molecular Programming Language
Natural Computing (2020) 19:391–407 DOI: 10.1007/s11047-019-09775-1
CRN++ on GitHub -
Inigo Quilez
https://www.iquilezles.org/www/index.htm
Introduction to Raymarching Signed Distance Functions


