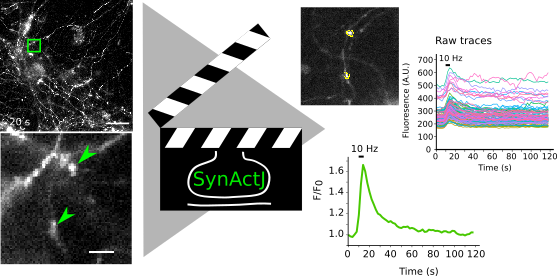
SynActJ is an image and data analysis workflow that allows to analyze synaptic activity. It is based on a Fiji plugin and a R Shiny App that implement the automated image analysis of active synapses in time-lapse movies. We tested the workflow with movies of pHluorin or calcium sensors.
This work has been published: https://doi.org/10.3389/fcomp.2021.777837
Please cite:
Schmied C, Soykan T, Bolz S, Haucke V and Lehmann M (2021) SynActJ: Easy-to-Use Automated Analysis of Synaptic Activity. Front. Comput. Sci. 3:777837. doi: 10.3389/fcomp.2021.777837
Have a look at the github pages site for more information:
https://schmiedc.github.io/SynActJ/
- Java Swing based graphical user interface
- Interactive adjustment over entire dataset
- Batch processing executed from main interface
- Saving and loading of processing settings
- Shiny App for data processing
Expected are 2D single channel .tif files containing multiple frames. At a specific frame the cultured neurons were stimulated and active boutons show an increase in intensity. The image calibration can be changed in the workflow. A settings file can be provided but can also be created later.
A small example file is provided here: Link to example data
The default segmentation parameters should work for this example file.
Tutorial for Shiny app: Link to tutorial
For the image analysis you need to download and install Fiji: Link to Fiji.
The plugin is available via an update site. Add the Cellular-Imaging site:
- Select Help › Update… from the menu bar. This will install potential updates and open a new window.
- Click on Manage update sites. Which opens the Manage update sites dialog.
- Search for the Cellular Imaging update site in the list.
- Add the update site by setting the tick box.
- Press Close and then Apply changes.
- The SynActJ should appear with the Status: Install it.
- Press Apply changes wait for download to finish and restart Fiji.
Repository for Shiny app: Link to Repo
Tutorial for Shiny app: Link to tutorial
For the data analysis you need to download R and RStudio: R Version 4.1.0
Link to R
Select version 4.1.0
RStudio 1.4.1717
Link to RStudio
- Download the contents of the repository: SynActJ Shiny
Click on the green button: Code.
Press Download ZIP to download the scripts. - Unzip the script to a location of your choice.
- Open the app.R file in RStudio.
- Start the application: press Run App - top right corner of RStudio.
- RStudio may ask to install or load extra packages - Download will take some time.
- Once these packages are installed and loaded the RShiny GUI should pop up.
- Optional: Press Open in Browser for a better rendering of the GUI.