This project is stable and being incubated for long-term support. It may contain new experimental code, for which APIs are subject to change.
Causal ML is a Python package that provides a suite of uplift modeling and causal inference methods using machine learning algorithms based on recent
research. It provides a standard interface that allows user to estimate the Conditional Average Treatment Effect (CATE) or Individual Treatment
Effect (ITE) from experimental or observational data. Essentially, it estimates the causal impact of intervention T on outcome Y for users
with observed features X, without strong assumptions on the model form. Typical use cases include
-
Campaign targeting optimization: An important lever to increase ROI in an advertising campaign is to target the ad to the set of customers who will have a favorable response in a given KPI such as engagement or sales. CATE identifies these customers by estimating the effect of the KPI from ad exposure at the individual level from A/B experiment or historical observational data.
-
Personalized engagement: A company has multiple options to interact with its customers such as different product choices in up-sell or messaging channels for communications. One can use CATE to estimate the heterogeneous treatment effect for each customer and treatment option combination for an optimal personalized recommendation system.
The package currently supports the following methods
- Tree-based algorithms
- Meta-learner algorithms
- Instrumental variables algorithms
- 2-Stage Least Squares (2SLS)
- Doubly Robust (DR) IV [8]
- Neural-network-based algorithms
Installation with conda is recommended. conda environment files for Python 3.6, 3.7 and 3.8 are available in the repository. Python 3.9 is not officially supported because some dependencies (e.g. tensorflow) does not support Python 3.9 yet.
This will create a new conda virtual environment named causalml-py36, causalml-py37, or causalml-py38. If you want to change the name of the environment, update envs/environment-py3x.yml.
$ git clone https://github.com/uber/causalml.git
$ cd causalml/envs/
$ conda env create -f environment-py38.yml # for the virtual environment with Python 3.8 and CausalML
$ conda activate causalml-py38
(causalml-py38)
$ git clone https://github.com/uber/causalml.git
$ cd causalml
$ pip install -r requirements.txt
$ pip install causalml
$ git clone https://github.com/uber/causalml.git
$ cd causalml
$ pip install -r requirements.txt
$ python setup.py build_ext --inplace
$ python setup.py install
from causalml.inference.meta import LRSRegressor
from causalml.inference.meta import XGBTRegressor, MLPTRegressor
from causalml.inference.meta import BaseXRegressor
from causalml.inference.meta import BaseRRegressor
from xgboost import XGBRegressor
from causalml.dataset import synthetic_data
y, X, treatment, _, _, e = synthetic_data(mode=1, n=1000, p=5, sigma=1.0)
lr = LRSRegressor()
te, lb, ub = lr.estimate_ate(X, treatment, y)
print('Average Treatment Effect (Linear Regression): {:.2f} ({:.2f}, {:.2f})'.format(te[0], lb[0], ub[0]))
xg = XGBTRegressor(random_state=42)
te, lb, ub = xg.estimate_ate(X, treatment, y)
print('Average Treatment Effect (XGBoost): {:.2f} ({:.2f}, {:.2f})'.format(te[0], lb[0], ub[0]))
nn = MLPTRegressor(hidden_layer_sizes=(10, 10),
learning_rate_init=.1,
early_stopping=True,
random_state=42)
te, lb, ub = nn.estimate_ate(X, treatment, y)
print('Average Treatment Effect (Neural Network (MLP)): {:.2f} ({:.2f}, {:.2f})'.format(te[0], lb[0], ub[0]))
xl = BaseXRegressor(learner=XGBRegressor(random_state=42))
te, lb, ub = xl.estimate_ate(X, e, treatment, y)
print('Average Treatment Effect (BaseXRegressor using XGBoost): {:.2f} ({:.2f}, {:.2f})'.format(te[0], lb[0], ub[0]))
rl = BaseRRegressor(learner=XGBRegressor(random_state=42))
te, lb, ub = rl.estimate_ate(X=X, p=e, treatment=treatment, y=y)
print('Average Treatment Effect (BaseRRegressor using XGBoost): {:.2f} ({:.2f}, {:.2f})'.format(te[0], lb[0], ub[0]))See the Meta-learner example notebook for details.
Causal ML provides methods to interpret the treatment effect models trained as follows:
from causalml.inference.meta import BaseSRegressor, BaseTRegressor, BaseXRegressor, BaseRRegressor
from causalml.dataset.regression import synthetic_data
# Load synthetic data
y, X, treatment, tau, b, e = synthetic_data(mode=1, n=10000, p=25, sigma=0.5)
w_multi = np.array(['treatment_A' if x==1 else 'control' for x in treatment]) # customize treatment/control names
slearner = BaseSRegressor(LGBMRegressor(), control_name='control')
slearner.estimate_ate(X, w_multi, y)
slearner_tau = slearner.fit_predict(X, w_multi, y)
model_tau_feature = RandomForestRegressor() # specify model for model_tau_feature
slearner.get_importance(X=X, tau=slearner_tau, model_tau_feature=model_tau_feature,
normalize=True, method='auto', features=feature_names)
# Using the feature_importances_ method in the base learner (LGBMRegressor() in this example)
slearner.plot_importance(X=X, tau=slearner_tau, normalize=True, method='auto')
# Using eli5's PermutationImportance
slearner.plot_importance(X=X, tau=slearner_tau, normalize=True, method='permutation')
# Using SHAP
shap_slearner = slearner.get_shap_values(X=X, tau=slearner_tau)
# Plot shap values without specifying shap_dict
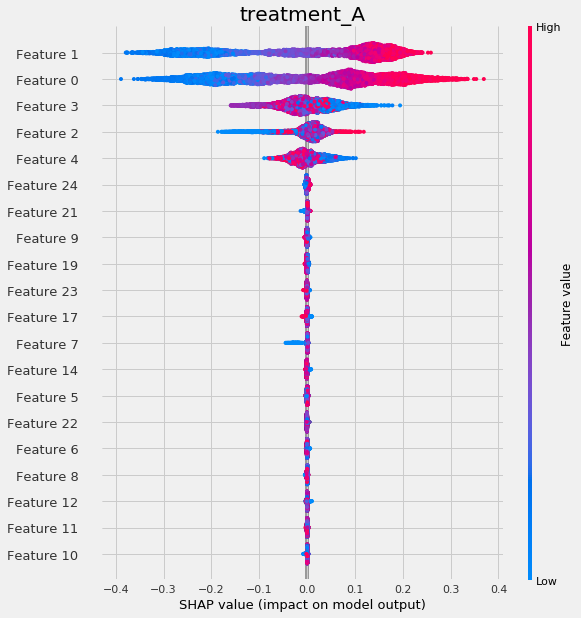
slearner.plot_shap_values(X=X, tau=slearner_tau)
# Plot shap values WITH specifying shap_dict
slearner.plot_shap_values(X=X, shap_dict=shap_slearner)
# interaction_idx set to 'auto' (searches for feature with greatest approximate interaction)
slearner.plot_shap_dependence(treatment_group='treatment_A',
feature_idx=1,
X=X,
tau=slearner_tau,
interaction_idx='auto')See the feature interpretations example notebook for details.
from IPython.display import Image
from causalml.inference.tree import UpliftTreeClassifier, UpliftRandomForestClassifier
from causalml.inference.tree import uplift_tree_string, uplift_tree_plot
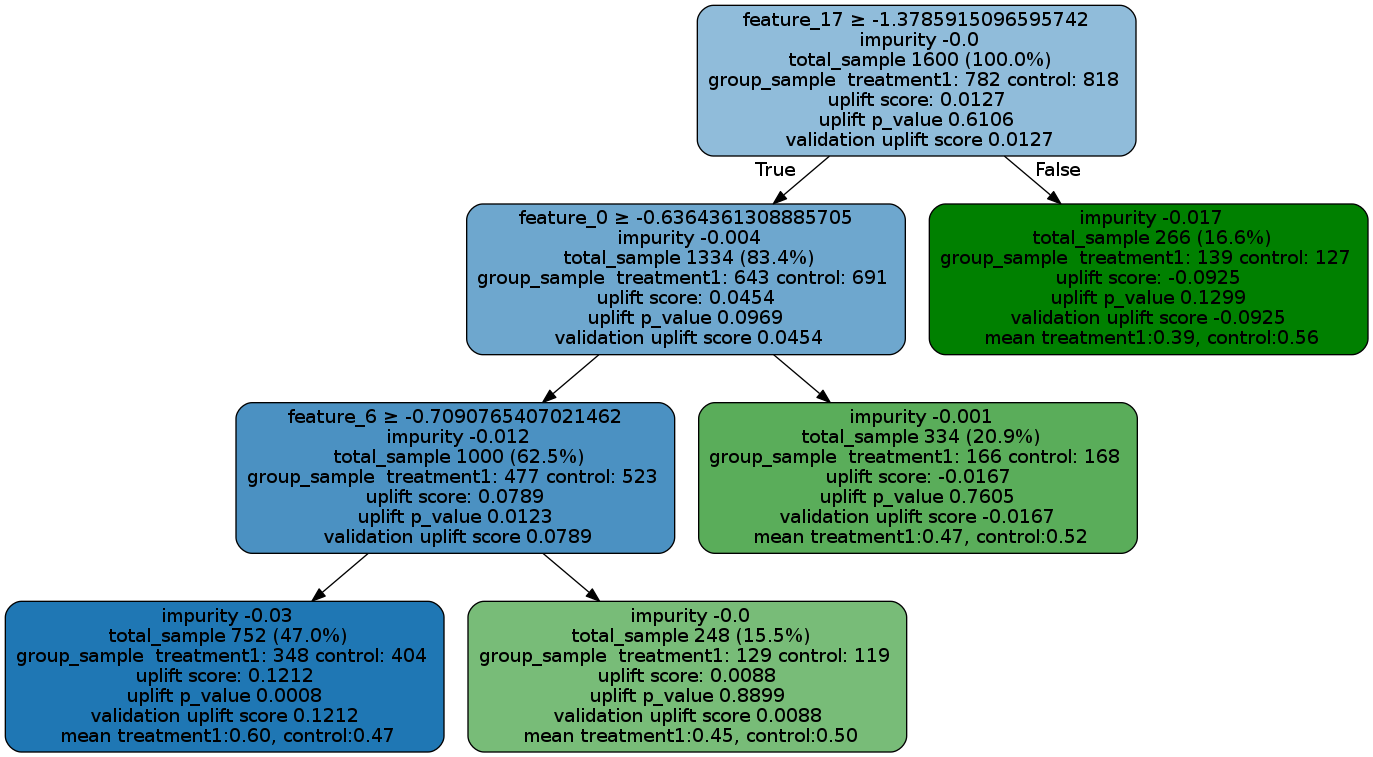
uplift_model = UpliftTreeClassifier(max_depth=5, min_samples_leaf=200, min_samples_treatment=50,
n_reg=100, evaluationFunction='KL', control_name='control')
uplift_model.fit(df[features].values,
treatment=df['treatment_group_key'].values,
y=df['conversion'].values)
graph = uplift_tree_plot(uplift_model.fitted_uplift_tree, features)
Image(graph.create_png())See the Uplift Tree visualization example notebook for details.
We welcome community contributors to the project. Before you start, please read our code of conduct and check out contributing guidelines first.
We document versions and changes in our changelog.
This project is licensed under the Apache 2.0 License - see the LICENSE file for details.
To cite CausalML in publications, you can refer to the following sources:
Whitepaper: CausalML: Python Package for Causal Machine Learning
Bibtex:
@misc{chen2020causalml, title={CausalML: Python Package for Causal Machine Learning}, author={Huigang Chen and Totte Harinen and Jeong-Yoon Lee and Mike Yung and Zhenyu Zhao}, year={2020}, eprint={2002.11631}, archivePrefix={arXiv}, primaryClass={cs.CY} }
- Radcliffe, Nicholas J., and Patrick D. Surry. "Real-world uplift modelling with significance-based uplift trees." White Paper TR-2011-1, Stochastic Solutions (2011): 1-33.
- Zhao, Yan, Xiao Fang, and David Simchi-Levi. "Uplift modeling with multiple treatments and general response types." Proceedings of the 2017 SIAM International Conference on Data Mining. Society for Industrial and Applied Mathematics, 2017.
- Athey, Susan, and Guido Imbens. "Recursive partitioning for heterogeneous causal effects." Proceedings of the National Academy of Sciences 113.27 (2016): 7353-7360.
- Künzel, Sören R., et al. "Metalearners for estimating heterogeneous treatment effects using machine learning." Proceedings of the national academy of sciences 116.10 (2019): 4156-4165.
- Nie, Xinkun, and Stefan Wager. "Quasi-oracle estimation of heterogeneous treatment effects." arXiv preprint arXiv:1712.04912 (2017).
- Bang, Heejung, and James M. Robins. "Doubly robust estimation in missing data and causal inference models." Biometrics 61.4 (2005): 962-973.
- Van Der Laan, Mark J., and Daniel Rubin. "Targeted maximum likelihood learning." The international journal of biostatistics 2.1 (2006).
- Kennedy, Edward H. "Optimal doubly robust estimation of heterogeneous causal effects." arXiv preprint arXiv:2004.14497 (2020).
- Louizos, Christos, et al. "Causal effect inference with deep latent-variable models." arXiv preprint arXiv:1705.08821 (2017).
- Shi, Claudia, David M. Blei, and Victor Veitch. "Adapting neural networks for the estimation of treatment effects." 33rd Conference on Neural Information Processing Systems (NeurIPS 2019), 2019.
- uplift: uplift models in R
- grf: generalized random forests that include heterogeneous treatment effect estimation in R
- rlearner: A R package that implements R-Learner
- DoWhy: Causal inference in Python based on Judea Pearl's do-calculus
- EconML: A Python package that implements heterogeneous treatment effect estimators from econometrics and machine learning methods